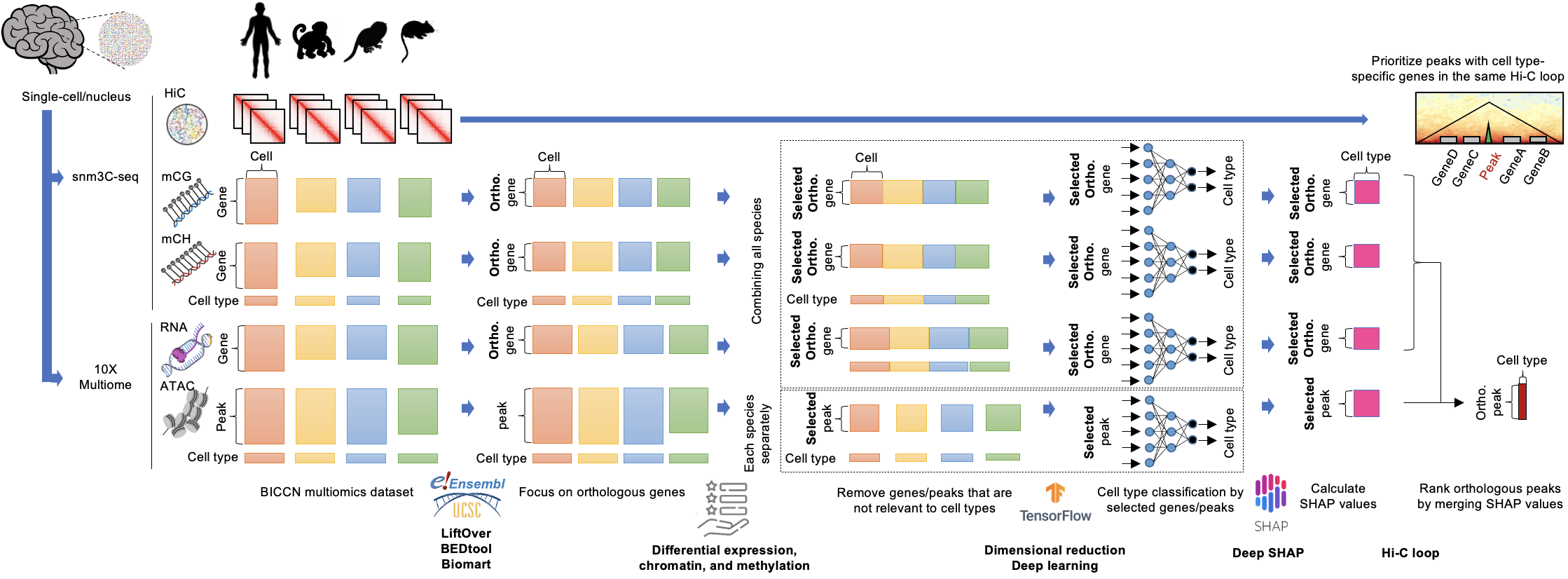
cisMultiDeep: Identifying Cell Type-Specific Cis-Regulatory Regions by Automatically-Tuned Deep Neural Network and SHAP
This repository presents the workflow to identify functional enhancers for each annotated cell type from a diverse collection of multi-omics profiles. The cell type is determined by expression of a specific set of genes that are governed by cis-regulatory elements in non-coding genomic regions. Recent studies have demonstrated that the cell type-specific CREs are not fully conserved (Villar et al., 2015), whereas the expression patterns of the cell type-specific genes are evolutionarily conserved (Shay et al., 2012). Here, we aim to identify the set of both ‘conserved’ and ‘non-conserved’ cell type-specific CREs that controls the expression of ‘conserved’ cell type-specific genes. cisMultiDeep employs automatically-tuned deep learning with SHAP feature importance assessment in cross-species single-cell multiomics data.
python = 3.10.2
R = 4.0.6
git clone https://github.com/ytanaka-bio/BICCN_Challenge_2023
cd BICCN_Challenge_2023
pip install -r requirements.txt --user
1.1. Download cross-species' multi-omics datasets that were provided from BICCN committee using AWS CLI as follow:
aws s3 sync s3://biccn-challenge . --no-sign-request
1.2. Add "chr" into Macaque bedpe files:
mkdir snm3C/Macaque2
mkdir snm3C/Macaque2/HiC_Loops
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Astro.loop.bedpe > snm3C/Macaque2/HiC_Loops/Astro.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/CLA.loop.bedpe > snm3C/Macaque2/HiC_Loops/CLA.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L23.loop.bedpe > snm3C/Macaque2/HiC_Loops/L23.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L4.loop.bedpe > snm3C/Macaque2/HiC_Loops/L4.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L5-ET.loop.bedpe > snm3C/Macaque2/HiC_Loops/L5-ET.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L5-IT.loop.bedpe > snm3C/Macaque2/HiC_Loops/L5-IT.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L6b.loop.bedpe > snm3C/Macaque2/HiC_Loops/L6b.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L6-CT.loop.bedpe > snm3C/Macaque2/HiC_Loops/L6-CT.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/L6-IT.loop.bedpe > snm3C/Macaque2/HiC_Loops/L6-IT.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Lamp5.loop.bedpe > snm3C/Macaque2/HiC_Loops/Lamp5.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/MG.loop.bedpe > snm3C/Macaque2/HiC_Loops/MG.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/NP.loop.bedpe > snm3C/Macaque2/HiC_Loops/NP.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/ODC.loop.bedpe > snm3C/Macaque2/HiC_Loops/ODC.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/OPC.loop.bedpe > snm3C/Macaque2/HiC_Loops/OPC.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Pvalb-BC.loop.bedpe > snm3C/Macaque2/HiC_Loops/Pvalb-BC.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Pvalb-ChC.loop.bedpe > snm3C/Macaque2/HiC_Loops/Pvalb-ChC.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Sncg.loop.bedpe > snm3C/Macaque2/HiC_Loops/Sncg.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Sst.loop.bedpe > snm3C/Macaque2/HiC_Loops/Sst.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Vip.loop.bedpe > snm3C/Macaque2/HiC_Loops/Vip.loop.bedpe
awk 'OFS="\t" {$1="chr"$1; $4="chr"$4; print}' snm3C/Macaque/HiC_Loops/Vsc.loop.bedpe > snm3C/Macaque2/HiC_Loops/Vsc.loop.bedpe
1.3. Download gene coordinate GTF files as follow:
#Human
wget https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_33/gencode.v33.annotation.gtf.gz
zcat gencode.v33.annotation.gtf.gz | grep gene_name | awk 'OFS="\t" {if ($3=="gene") {print $1,$4-1,$5,$14,$7}}' | tr -d '";' | sort -k1,1 -k2,2n -k3,3n > Human_gene.bed
mv gencode.v33.annotation.gtf.gz Human_gene.gtf.gz
#Macaque
wget https://ftp.ensembl.org/pub/release-110/gtf/macaca_mulatta/Macaca_mulatta.Mmul_10.110.gtf.gz
zcat Macaca_mulatta.Mmul_10.110.gtf.gz | sed -E 's/^(\w+)/chr\1/g' > Macaque_gene.gtf
rm Macaca_mulatta.Mmul_10.110.gtf.gz
grep gene_name Macaque_gene.gtf | awk 'OFS="\t" {if ($3=="gene") {print $1,$4-1,$5,$14,$7}}' | tr -d '";' | sort -k1,1 -k2,2n -k3,3n > Macaque_gene.bed
gzip Macaque_gene.gtf
#Marmoset
wget https://hgdownload.soe.ucsc.edu/goldenPath/calJac4/bigZips/genes/ncbiRefSeq.gtf.gz
zcat ncbiRefSeq.gtf.gz | grep gene_name | awk 'OFS="\t" {if ($3=="transcript") {print $1,$4-1,$5,$14,$7}}' | tr -d '";' | sort -k1,1 -k2,2n -k3,3n > Marmoset_gene.bed
mv ncbiRefSeq.gtf.gz Marmoset_gene.gtf.gz
#Mouse
wget ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M22/gencode.vM22.annotation.gtf.gz
zcat gencode.vM22.annotation.gtf.gz | grep gene_name | awk 'OFS="\t" {if ($3=="gene") {print $1,$4-1,$5,$14,$7}}' | tr -d '";' | sort -k1,1 -k2,2n -k3,3n > Mouse_gene.bed
mv gencode.vM22.annotation.gtf.gz Mouse_gene.gtf.gz
1.4. Remove duplicated genes from BED file:
R CMD BATCH remove_duplicate_gene_bed.R
1.5. Obtain orthologous gene list from Biomart as follow:
- Choose "Ensembl Gene 10" and "Human genes (GRCh39.p14)"
- In Attributes section, choose "Homologues (Max select 6 orthologues)"
- In GENE tab, choose "Gene stable ID" and "Gene name"
- In ORTHOLOGUES [K-O] tab, choose "Macaque gene table ID", "Macaque gene name", "Mouse gene stable ID", and "Mouse gene name"
- In ORTHOLOGUES [U-Z] tab, choose "White-tufted-ear marmoset gene stable ID" and "White-tufted-ear marmoset gene name"
- Click "Result"
- Choose "compressed file (.gz)" in export all results, and click "GO"
2.1. Calculate the cell type specificity score for each gene from transcriptome (RNA):
python identify_celltype_gene.py
2.2. Calculate the cell type specificity score for each gene from methylome (mCG, mCH):
python identify_celltype_methyl.py
2.3. Calculate the cell type specificity score for each peak from chromatin accessibility profiles (ATAC):
python identify_celltype_peak.py
3.1. Download LiftOver UCSC Chain files:
wget https://hgdownload.soe.ucsc.edu/goldenPath/mm10/liftOver/mm10ToHg38.over.chain.gz #Mouse vs Human
wget https://hgdownload.soe.ucsc.edu/goldenPath/mm10/liftOver/mm10ToRheMac10.over.chain.gz #Mouse vs Macaque
wget https://hgdownload.soe.ucsc.edu/goldenPath/mm10/liftOver/mm10ToCalJac4.over.chain.gz #Mouse vs Marmoset
3.2. Generate BED format files of ATAC peaks:
R CMD BATCH get_atac_bed.R
3.3. Run LiftOver command to identify orthologous regions of Mouse ATAC peaks in other species' genomes:
liftOver Mouse_atac.bed mm10ToHg38.over.chain.gz Mouse_atac_Human.bed Mouse_atac_Human_unmapped.bed
liftOver Mouse_atac.bed mm10ToRheMac10.over.chain.gz Mouse_atac_Macaque.bed Mouse_atac_Macaque_unmapped.bed
liftOver Mouse_atac.bed mm10ToCalJac4.over.chain.gz Mouse_atac_Marmoset.bed Mouse_atac_Marmoset_unmapped.bed
3.4. Run intersect command of BEDTools to identify ATAC peaks in each species corresponding to Mouse ATAC peaks:
bedtools intersect -a Mouse_atac_Human.bed -b Human_atac.bed -wa -wb -f 0.5 > Mouse_atac_Human_overlap.bed
bedtools intersect -a Mouse_atac_Macaque.bed -b Macaque_atac.bed -wa -wb -f 0.5 > Mouse_atac_Macaque_overlap.bed
bedtools intersect -a Mouse_atac_Marmoset.bed -b Marmoset_atac.bed -wa -wb -f 0.5 > Mouse_atac_Marmoset_overlap.bed
4.1. Get conserved gene/peak list using a R script get_cons_data.R:
R CMD BATCH get_cons_data.R
4.2. Prepare input and output dataset for Deep Learning (Here, we focus on top 1000/10000 differential genes/peaks in each cell type):
#RNA, mCG, mCH
python prepare_cons_dataset.py -f 10XMultiome/Mouse/Mouse_rna.h5ad 10XMultiome/Human/Human_rna.h5ad 10XMultiome/Macaque/Macaque_rna.h5ad 10XMultiome/Marmoset/Marmoset_rna.h5ad -d all_rna_dif_cons.csv -a subclass_Bakken_2022 -r cons_rna_list.csv -o rna_1000 -g 1000
python prepare_cons_dataset.py -f snm3C/Mouse/Mouse_mCG_gene_fractions.h5ad snm3C/Human/Human_mCG_gene_fractions.h5ad snm3C/Macaque/Macaque_mCG_gene_fractions.h5ad snm3C/Marmoset/Marmoset_mCG_gene_fractions.h5ad -d all_mCG_dif_cons.csv -a subclass_Bakken_2022 -r cons_mCG_list.csv -o mCG_1000 -g 1000 -n False
python prepare_cons_dataset.py -f snm3C/Mouse/Mouse_mCH_gene_fractions.h5ad snm3C/Human/Human_mCH_gene_fractions.h5ad snm3C/Macaque/Maacque_mCH_gene_fractions.h5ad snm3C/Marmoset/Marmoset_mCH_gene_fractions.h5ad -d all_mCH_dif_cons.csv -a subclass_Bakken_2022 -r cons_mCH_list.csv -o mCH_1000 -g 1000 -n False
#ATAC
python prepare_dataset.py -f 10XMultiome/Mouse/Mouse_atac.h5ad -d Mouse_atac_dif_selected.csv -a subclass_Bakken_2022 -o Mouse_atac_10000 -g 10000
python prepare_dataset.py -f 10XMultiome/Human/Human_atac.h5ad -d Human_atac_dif_selected.csv -a subclass_Bakken_2022 -o Human_atac_10000 -g 10000
python prepare_dataset.py -f 10XMultiome/Macaque/Macaque_atac.h5ad -d Macaque_atac_dif_selected.csv -a subclass_Bakken_2022 -o Macaque_atac_10000 -g 10000
python prepare_dataset.py -f 10XMultiome/Marmoset/Marmoset_atac.h5ad -d Marmoset_atac_dif_selected.csv -a subclass_Bakken_2022 -o Marmoset_atac_10000 -g 10000
5.1. Train Deep learning model and calculate the contribution (SHAP value) of each gene/peak to the segregation of cell types:
#RNA, mCG, mCH of conserved genes
python DeepSHAP_sparse.py -i rna_1000 -p rna_1000_output.csv -o rna_1000_deep_DR -t 12
python DeepSHAP_sparse.py -i mCG_1000 -p mCG_1000_output.csv -o mCG_1000_deep_DR -t 12
python DeepSHAP_sparse.py -i mCH_1000 -p mCH_1000_output.csv -o mCH_1000_deep_DR -t 12
#Mouse ATAC peaks
python DeepSHAP_sparse.py -i Mouse_atac_10000 -p Mouse_atac_10000_output.csv -o Mouse_atac_10000_deep_DR -t 12
python DeepSHAP_sparse.py -i Human_atac_10000 -p Human_atac_10000_output.csv -o Human_atac_10000_deep_DR -t 12
python DeepSHAP_sparse.py -i Macaque_atac_10000 -p Macaque_atac_10000_output.csv -o Macaque_atac_10000_deep_DR -t 12
python DeepSHAP_sparse.py -i Marmoset_atac_10000 -p Marmoset_atac_10000_output.csv -o Marmoset_atac_10000_deep_DR -t 12
6.1. Prepare gene and peak BED file for differential genes/peaks:
R CMD BATCH prepare_bed.R
6.2. Identify genes and peaks within each HiC loop:
mkdir inLoop
#Human ATAC
bedtools pairtobed -a snm3C/Human/HiC_Loops/Astro.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Astro.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/CLA.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_CLA.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L23.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_L23.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-ET.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_L5-ET.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-IT.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_L5-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/NP.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_NP.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-CT.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_L6-CT.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-IT.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_L6-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6b.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_L6b.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Lamp5.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Lamp5.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/MG.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_MG.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/ODC.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_ODC.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/OPC.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_OPC.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Pvalb-BC.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Pvalb-BC.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sncg.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Sncg.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sst.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Sst.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vip.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Vip.loop_atac.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vsc.loop.bedpe -b Human_atac_10000.bed > inLoop/Human_Vsc.loop_atac.txt
#Human RNA
bedtools pairtobed -a snm3C/Human/HiC_Loops/Astro.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Astro.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/CLA.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_CLA.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L23.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_L23.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-ET.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_L5-ET.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-IT.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_L5-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/NP.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_NP.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-CT.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_L6-CT.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-IT.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_L6-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6b.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_L6b.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Lamp5.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Lamp5.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/MG.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_MG.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/ODC.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_ODC.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/OPC.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_OPC.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Pvalb-BC.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Pvalb-BC.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sncg.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Sncg.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sst.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Sst.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vip.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Vip.loop_rna.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vsc.loop.bedpe -b Human_rna_1000.bed > inLoop/Human_Vsc.loop_rna.txt
#Human mCG
bedtools pairtobed -a snm3C/Human/HiC_Loops/Astro.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Astro.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/CLA.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_CLA.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L23.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_L23.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-ET.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_L5-ET.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-IT.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_L5-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/NP.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_NP.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-CT.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_L6-CT.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-IT.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_L6-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6b.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_L6b.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Lamp5.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Lamp5.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/MG.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_MG.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/ODC.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_ODC.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/OPC.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_OPC.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Pvalb-BC.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Pvalb-BC.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sncg.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Sncg.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sst.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Sst.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vip.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Vip.loop_mCG.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vsc.loop.bedpe -b Human_mCG_1000.bed > inLoop/Human_Vsc.loop_mCG.txt
#Human mCH
bedtools pairtobed -a snm3C/Human/HiC_Loops/Astro.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Astro.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/CLA.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_CLA.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L23.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_L23.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-ET.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_L5-ET.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L5-IT.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_L5-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/NP.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_NP.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-CT.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_L6-CT.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6-IT.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_L6-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/L6b.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_L6b.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Lamp5.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Lamp5.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/MG.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_MG.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/ODC.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_ODC.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/OPC.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_OPC.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Pvalb-BC.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Pvalb-BC.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sncg.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Sncg.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Sst.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Sst.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vip.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Vip.loop_mCH.txt
bedtools pairtobed -a snm3C/Human/HiC_Loops/Vsc.loop.bedpe -b Human_mCH_1000.bed > inLoop/Human_Vsc.loop_mCH.txt
#Macaque ATAC
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Astro.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Astro.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/CLA.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_CLA.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L23.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_L23.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-ET.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_L5-ET.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-IT.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_L5-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/NP.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_NP.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-CT.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_L6-CT.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-IT.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_L6-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6b.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_L6b.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Lamp5.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Lamp5.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/MG.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_MG.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/ODC.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_ODC.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/OPC.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_OPC.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Pvalb-BC.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Pvalb-BC.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sncg.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Sncg.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sst.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Sst.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vip.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Vip.loop_atac.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vsc.loop.bedpe -b Macaque_atac_10000.bed > inLoop/Macaque_Vsc.loop_atac.txt
#Macaque RNA
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Astro.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Astro.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/CLA.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_CLA.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L23.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_L23.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-ET.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_L5-ET.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-IT.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_L5-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/NP.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_NP.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-CT.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_L6-CT.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-IT.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_L6-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6b.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_L6b.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Lamp5.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Lamp5.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/MG.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_MG.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/ODC.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_ODC.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/OPC.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_OPC.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Pvalb-BC.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Pvalb-BC.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sncg.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Sncg.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sst.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Sst.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vip.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Vip.loop_rna.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vsc.loop.bedpe -b Macaque_rna_1000.bed > inLoop/Macaque_Vsc.loop_rna.txt
#Macaque mCG
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Astro.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Astro.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/CLA.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_CLA.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L23.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_L23.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-ET.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_L5-ET.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-IT.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_L5-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/NP.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_NP.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-CT.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_L6-CT.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-IT.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_L6-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6b.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_L6b.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Lamp5.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Lamp5.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/MG.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_MG.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/ODC.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_ODC.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/OPC.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_OPC.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Pvalb-BC.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Pvalb-BC.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sncg.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Sncg.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sst.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Sst.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vip.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Vip.loop_mCG.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vsc.loop.bedpe -b Macaque_mCG_1000.bed > inLoop/Macaque_Vsc.loop_mCG.txt
#Macaque mCH
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Astro.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Astro.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/CLA.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_CLA.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L23.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_L23.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-ET.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_L5-ET.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L5-IT.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_L5-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/NP.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_NP.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-CT.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_L6-CT.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6-IT.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_L6-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/L6b.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_L6b.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Lamp5.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Lamp5.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/MG.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_MG.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/ODC.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_ODC.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/OPC.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_OPC.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Pvalb-BC.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Pvalb-BC.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sncg.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Sncg.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Sst.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Sst.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vip.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Vip.loop_mCH.txt
bedtools pairtobed -a snm3C/Macaque2/HiC_Loops/Vsc.loop.bedpe -b Macaque_mCH_1000.bed > inLoop/Macaque_Vsc.loop_mCH.txt
#Marmoset ATAC
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Astro.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Astro.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/CLA.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_CLA.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L23.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_L23.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-ET.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_L5-ET.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-IT.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_L5-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/NP.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_NP.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-CT.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_L6-CT.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-IT.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_L6-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6b.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_L6b.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Lamp5.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Lamp5.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/MG.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_MG.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/ODC.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_ODC.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/OPC.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_OPC.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Pvalb-BC.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Pvalb-BC.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sncg.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Sncg.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sst.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Sst.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vip.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Vip.loop_atac.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vsc.loop.bedpe -b Marmoset_atac_10000.bed > inLoop/Marmoset_Vsc.loop_atac.txt
#Marmoset RNA
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Astro.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Astro.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/CLA.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_CLA.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L23.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_L23.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-ET.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_L5-ET.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-IT.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_L5-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/NP.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_NP.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-CT.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_L6-CT.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-IT.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_L6-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6b.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_L6b.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Lamp5.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Lamp5.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/MG.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_MG.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/ODC.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_ODC.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/OPC.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_OPC.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Pvalb-BC.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Pvalb-BC.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sncg.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Sncg.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sst.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Sst.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vip.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Vip.loop_rna.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vsc.loop.bedpe -b Marmoset_rna_1000.bed > inLoop/Marmoset_Vsc.loop_rna.txt
#Marmoset mCG
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Astro.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Astro.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/CLA.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_CLA.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L23.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_L23.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-ET.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_L5-ET.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-IT.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_L5-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/NP.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_NP.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-CT.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_L6-CT.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-IT.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_L6-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6b.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_L6b.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Lamp5.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Lamp5.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/MG.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_MG.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/ODC.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_ODC.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/OPC.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_OPC.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Pvalb-BC.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Pvalb-BC.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sncg.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Sncg.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sst.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Sst.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vip.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Vip.loop_mCG.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vsc.loop.bedpe -b Marmoset_mCG_1000.bed > inLoop/Marmoset_Vsc.loop_mCG.txt
#Marmoset mCH
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Astro.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Astro.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/CLA.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_CLA.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L23.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_L23.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-ET.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_L5-ET.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L5-IT.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_L5-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/NP.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_NP.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-CT.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_L6-CT.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6-IT.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_L6-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/L6b.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_L6b.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Lamp5.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Lamp5.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/MG.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_MG.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/ODC.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_ODC.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/OPC.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_OPC.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Pvalb-BC.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Pvalb-BC.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sncg.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Sncg.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Sst.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Sst.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vip.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Vip.loop_mCH.txt
bedtools pairtobed -a snm3C/Marmoset/HiC_Loops/Vsc.loop.bedpe -b Marmoset_mCH_1000.bed > inLoop/Marmoset_Vsc.loop_mCH.txt
#Mouse ATAC
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Astro.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Astro.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/CLA.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_CLA.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L23.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_L23.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-ET.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_L5-ET.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-IT.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_L5-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/NP.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_NP.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-CT.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_L6-CT.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-IT.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_L6-IT.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6b.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_L6b.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Lamp5.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Lamp5.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/MG.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_MG.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/ODC.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_ODC.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/OPC.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_OPC.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Pvalb-BC.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Pvalb-BC.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sncg.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Sncg.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sst.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Sst.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vip.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Vip.loop_atac.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vsc.loop.bedpe -b Mouse_atac_10000.bed > inLoop/Mouse_Vsc.loop_atac.txt
#Mouse RNA
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Astro.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Astro.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/CLA.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_CLA.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L23.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_L23.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-ET.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_L5-ET.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-IT.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_L5-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/NP.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_NP.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-CT.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_L6-CT.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-IT.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_L6-IT.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6b.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_L6b.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Lamp5.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Lamp5.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/MG.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_MG.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/ODC.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_ODC.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/OPC.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_OPC.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Pvalb-BC.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Pvalb-BC.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sncg.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Sncg.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sst.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Sst.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vip.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Vip.loop_rna.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vsc.loop.bedpe -b Mouse_rna_1000.bed > inLoop/Mouse_Vsc.loop_rna.txt
#Mouse mCG
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Astro.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Astro.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/CLA.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_CLA.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L23.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_L23.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-ET.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_L5-ET.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-IT.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_L5-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/NP.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_NP.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-CT.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_L6-CT.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-IT.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_L6-IT.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6b.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_L6b.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Lamp5.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Lamp5.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/MG.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_MG.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/ODC.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_ODC.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/OPC.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_OPC.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Pvalb-BC.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Pvalb-BC.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sncg.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Sncg.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sst.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Sst.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vip.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Vip.loop_mCG.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vsc.loop.bedpe -b Mouse_mCG_1000.bed > inLoop/Mouse_Vsc.loop_mCG.txt
#Mouse mCH
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Astro.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Astro.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/CLA.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_CLA.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L23.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_L23.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-ET.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_L5-ET.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L5-IT.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_L5-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/NP.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_NP.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-CT.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_L6-CT.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6-IT.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_L6-IT.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/L6b.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_L6b.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Lamp5.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Lamp5.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/MG.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_MG.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/ODC.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_ODC.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/OPC.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_OPC.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Pvalb-BC.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Pvalb-BC.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sncg.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Sncg.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Sst.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Sst.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vip.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Vip.loop_mCH.txt
bedtools pairtobed -a snm3C/Mouse/HiC_Loops/Vsc.loop.bedpe -b Mouse_mCH_1000.bed > inLoop/Mouse_Vsc.loop_mCH.txt
6.3. Calculate the sum of SHAP values of the connected genes by HiC loops.
R CMD BATCH all_sum_SHAP.R