spikesort is a framework written in MATLAB to help you sort spikes from single-channel extracellular recordings. It is
- highly modular Almost everything is written as a plugin.
spikesortis a MATLAB class, and plugins are methods defined within the class. - Accurate Using the t-SNE dimensionality reduction algorithm,
spikesortachieves a 99.5% accuracy on test data. For a full report, see this. - Data-agnostic
spikesortinterfaces to your data through plugins, andspikesortdoesn't care what your data format is. - Bring-your-own-algorithm
spikesortsplits up the spike sorting problem into two steps: dimensionality reduction and clustering. Every algorithm in either step is written as a plugin, and you can write your and drop it in, with zero modifications to the core code. For example,spikesortcan use the amazing mutli-core t-SNE algorithm to embed spike shapes in two dimensions very rapidly.
spikesort is written in MATLAB, with a sprinkling of Python wrapper code. It should work on any OS that modern MATLAB runs on, but has only been tested on macOS Sierra with MATLAB R2015a.
The best way to install spikesort is through my package manager:
% copy and paste this code in your MATLAB prompt
websave('install.m','https://git.io/viWvp')
install -f sg-s/spikesort
install -f sg-s/srinivas.gs_mtools % spikesort needs this package to run
This script grabs the code and fixes your path.
Or, if you have git installed:
git clone git@github.com:sg-s/spikesort.git
Don't forget to download, install and configure the other packages too (see below).
install bhtsne
If you used install.m to install spikesort, you already have these toolbox. However, you need to compile to run. On *nix-like systems, navigate to the folder it is in (should be ~/code/bhtsne/) and run the following:
g++ sptree.cpp tsne.cpp -o bh_tsne -O2
Note that my fork of bhtsne may have modifications, and this is what you should use with spikesort. If you're running Windows, you need to use whatever compiler you have to compile this. See this for instructions on how to build the binary.
install Multicore-TSNE
This implementation of t-SNE is the the fastest I know of, and gets faster the more cores you throw at it. Follow the installation instructions here. macOS users, see this for installation troubleshooting.
You then need to tell MATLAB where Multicore-TSNE is installed. To do this,
- first determine where it is installed (
pip show MulticoreTSNE) - Copy and paste this code to your MATLAB
startup.m(useedit startup.m)
a_path = '/path/to/MulticoreTSNE';
path1 = getenv('PATH');
if isempty(strfind(path1,[pathsep a_path]))
path1 = [a_path pathsep path1];
end
setenv('PATH', path1);- sorting into only two neurons/units (A and B) is supported. spikesort will not support more than 2 units in the anticipated future.
- Only 1 recording electrode is supported at a time. No support for multi-electrode arrays, nor will
spikesortever have support for MEAs.
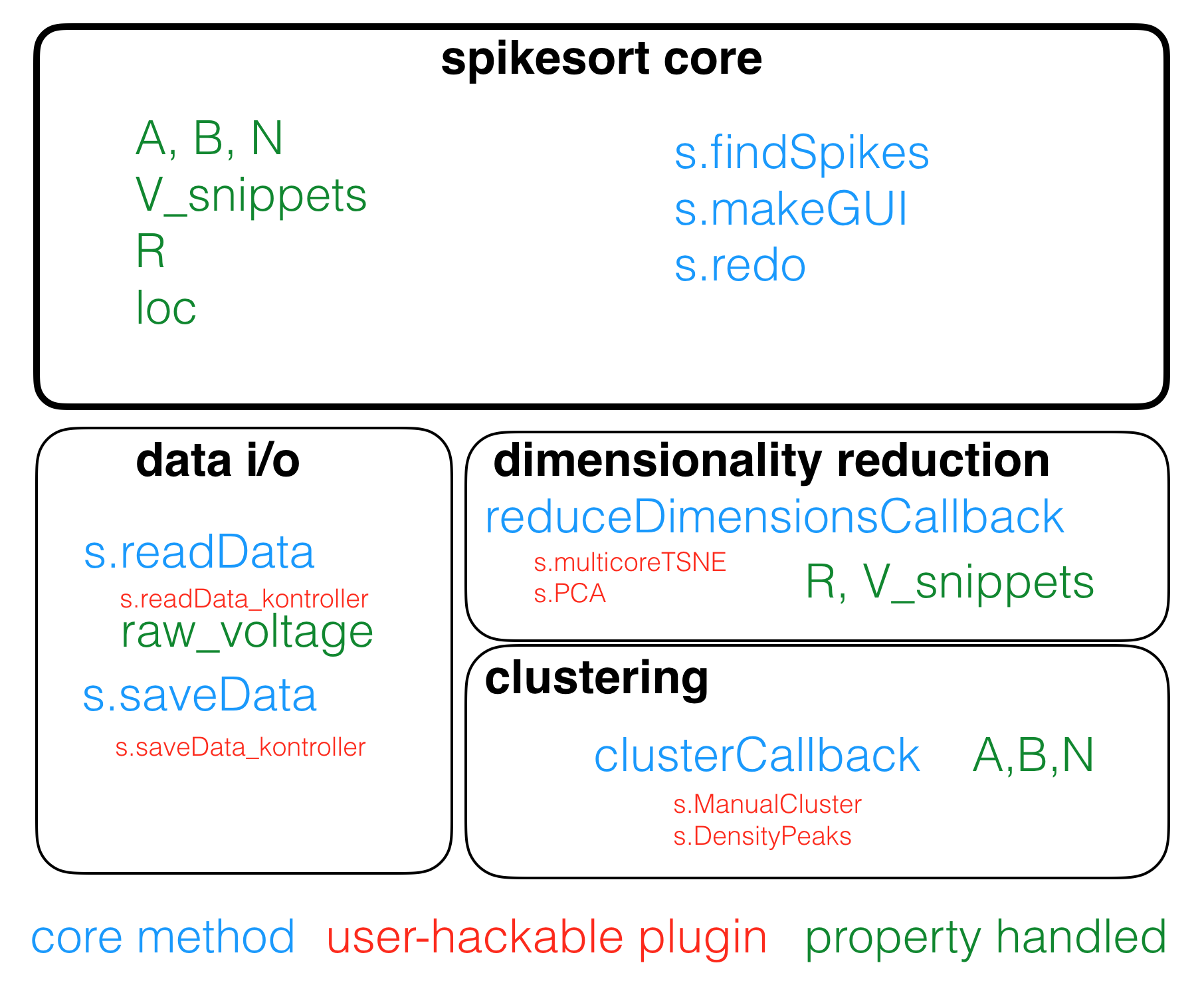
spikesort is built around a plugin architecture for the three most important things it does:
- Data handling
- Dimensionality reduction of spike shapes
- Clustering
Writing your own plugins is really easy: plugins are methods that you can simply drop into the spikesort classdef folder (@spikesort), and spikesort automatically figures out which methods are plugins (see naming convention below)
Plugins can be named whatever you want, though you are encouraged to use camelCase for all methods. The first three lines of every plugin should conform to the following convention:
% spikesort plugin
% plugin_type = 'dim-red';
% plugin_dimension = 2;
%
The first line identifies the method as a spikesort plugin, and the second line determines the type of plugin it is. Currently, plugins can be of six types:
dim-redclusterread-datasave-dataload-fileplot
If you are writing a read-data, save-data,load-file or plot plugin, the convention for the first three lines is as follows:
% spikesort plugin
% plugin_type = 'load-file';
% data_extension = 'kontroller'
% data_extension identifies the extension that spikesort binds that plugin to.
load-file plugins are expected to populate the following fields in the spikesort object:
output_channel_names
sampling_rate
this_trial
this_paradigm
handles.paradigm_chooser.String
plot-spikes plugins are expected to read all spikes in that data file, and make a raster or a firing rate plot, with appropriate labels for each trial and paradigm set.
If you plan to use spikesort for a publication, please write to me for appropriate citation.