Export MS Annika crosslink results to different file formats for different down-stream analysis tools.
Python 3.7+ installation with pandas, openpyxl, biopython and biopandas to run the scripts or to use the Proteome Discoverer Scripting Nodes.
- Install pandas:
pip install pandas - Install openpyxl:
pip install openpyxl - Install biopython:
pip install biopython - Install biopandas:
pip install biopandas
Alternatively there are Windows binaries available in the Releases tab that don't require a python installation.
FASTA headers need to follow the UniProtKB standard formatting (as described here) otherwise scripts may not work properly. The minimal requirement for FASTA headers is db|identifier|entry.
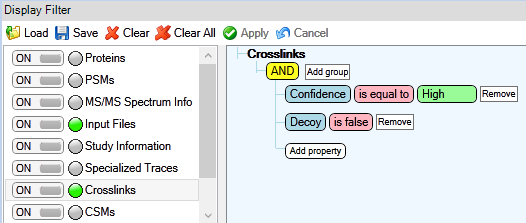
All of the scripts use Micrsoft Excel files as input, for that MS Annika results need to be exported from Proteome Discoverer. It is recommended to first filter results according to your needs, e.g. filter for high-confidence crosslinks and filter out decoy crosslinks as depicted below.
Figure 1: Crosslinks filtered for 1% estimated FDR and without decoys.
Results can then be exported by selecting File > Export > To Microsoft Excel… > Level 1: Crosslinks > Export in Proteome Discoverer.
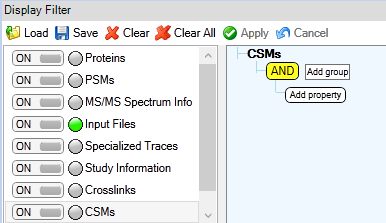
Figure 2: All (unvalidated) CSMs.
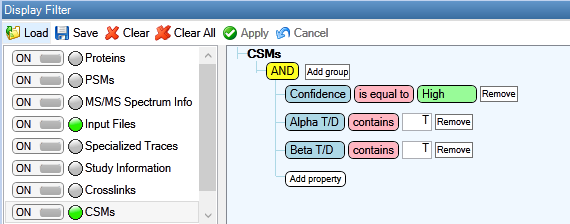
Figure 3: CSMs filtered for 1% estimated FDR and without decoys.
Results can then be exported by selecting File > Export > To Microsoft Excel… > Level 1: CSMs > Export in Proteome Discoverer.
- Exporting to xiNET
Files needed:- result.xlsx - MS Annika crosslink result file(s) exported to .xlsx
- seq.fasta - FASTA file containing sequences of the crosslinked proteins
python xiNetExporter_msannika.py result.xlsx -fasta seq.fasta - Exporting to xiVIEW
Files needed:- result.xlsx - MS Annika crosslink result file(s) exported to .xlsx
- seq.fasta - FASTA file containing sequences of the crosslinked proteins
python xiViewExporter_msannika.py result.xlsx -fasta seq.fasta - Exporting to xiFDR
Files needed (requires MS Annika 3.0.2 or later):- result.xlsx - MS Annika CSM result file (unvalidated) exported to .xlsx
# for xiFDR version < 2.2.1 python xiFdrExporter_msannika.py result.xlsx # for xiFDR version >= 2.2.1 python xiFdr2.2.1Exporter_msannika.py result.xlsx - Exporting to pyXlinkViewer (pyMOL)
Files needed:- result.xlsx - MS Annika crosslink result file(s) exported to .xlsx
- structure.pdb - 3D structure of the protein (complex) that crosslinks should be mapped to, alternatively you can also just provide the 4-letter code from the PDB and the script will fetch the structure from internet
python pyXlinkViewerExporter_msannika.py result.xlsx -pdb structure.pdb - Exporting to XLMS-Tools
XLMS-Tools uses the same file format as pyXlinkViewer, therefore the same exporter can be used! - Exporting to XMAS (ChimeraX)
Visualization of MS Annika results works out of the box with .xlsx files exported from Proteome Discoverer. - Exporting to PAE Viewer
Files needed:- pyXlinkViewer_export.csv - Crosslinks exported from pyXlinkViewer as .csv
python PAEViewerExporter_msannika.py pyXlinkViewer_export.csv
Export to xiNET
EXPORTER DESCRIPTION:
A script to export MS Annika results to xiNET input files (CSV + FASTA).
USAGE:
xiNetExporter_msannika.py f [f ...]
[-fasta FASTA]
[-ignore IGNORE]
[-o OUTPUT]
[-h]
[--version]
positional arguments:
f MS Annika crosslink result files in Microsoft Excel
format (.xlsx) to process.
required arguments:
-fasta FASTAFILE, --fasta FASTAFILE
Fasta file used for crosslink search. Must contain
proteins identified in the MS Annika result files.
optional arguments:
-ignore ACCESSION, --ignore ACCESSION
Protein accessions to be ignored. Crosslinks that only
link between ignored proteins will not be exported.
Supports input of multiple accessions.
-h, --help show this help message and exit
-o OUTPUT, --output OUTPUT
Prefix of the output files.
--version show program's version number and exit
Example usage:
python xiNetExporter_msannika.py "202001216_nsp8_trypsin_XL_REP1.xlsx" "202001216_nsp8_trypsin_XL_REP2.xlsx" "202001216_nsp8_trypsin_XL_REP3.xlsx" --fasta SARS-COV-2.fasta -o test --ignore P0DTC1 P0DTD1 P0DTC2
Or using the Windows binary:
xiNetExporter_msannika.exe "202001216_nsp8_trypsin_XL_REP1.xlsx" "202001216_nsp8_trypsin_XL_REP2.xlsx" "202001216_nsp8_trypsin_XL_REP3.xlsx" --fasta SARS-COV-2.fasta -o test --ignore P0DTC1 P0DTD1 P0DTC2
Export to xiVIEW
EXPORTER DESCRIPTION:
A script to export MS Annika results to xiVIEW input files (CSV + FASTA).
USAGE:
xiViewExporter_msannika.py f [f ...]
[-fasta FASTA]
[-ignore IGNORE]
[-o OUTPUT]
[-h]
[--version]
positional arguments:
f MS Annika crosslink result files in Microsoft Excel
format (.xlsx) to process.
required arguments:
-fasta FASTAFILE, --fasta FASTAFILE
Fasta file used for crosslink search. Must contain
proteins identified in the MS Annika result files.
optional arguments:
-ignore ACCESSION, --ignore ACCESSION
Protein accessions to be ignored. Crosslinks that only
link between ignored proteins will not be exported.
Supports input of multiple accessions.
-h, --help show this help message and exit
-o OUTPUT, --output OUTPUT
Prefix of the output files.
--version show program's version number and exit
Example usage:
python xiViewExporter_msannika.py "202001216_nsp8_trypsin_XL_REP1.xlsx" "202001216_nsp8_trypsin_XL_REP2.xlsx" "202001216_nsp8_trypsin_XL_REP3.xlsx" --fasta SARS-COV-2.fasta -o test --ignore P0DTC1 P0DTD1 P0DTC2
Or using the Windows binary:
xiViewExporter_msannika.exe "202001216_nsp8_trypsin_XL_REP1.xlsx" "202001216_nsp8_trypsin_XL_REP2.xlsx" "202001216_nsp8_trypsin_XL_REP3.xlsx" --fasta SARS-COV-2.fasta -o test --ignore P0DTC1 P0DTD1 P0DTC2
Export to xiFDR
Exporting to and validation with xiFDR requires MS Annika 3.0.2 or later!
EXPORTER DESCRIPTION:
A script to export MS Annika CSM results (.xlsx) to a xiFDR input file (.csv).
CSMs should be unfiltered, therefore include decoys and not be validated for any
FDR.
Warning: Exporter for xiFDR < 2.2.1 currently only reports one/the first protein for
ambiguous peptides that are found in more than one protein!
USAGE:
xiFdrExporter_msannika.py f [f]
[-o OUTPUT]
[-h]
[--version]
positional arguments:
f Crosslink-Spectrum-Matches (CSMs) exported from
MS Annika in Microsoft Excel (.xlsx) format.
optional arguments:
-o OUTPUT, --output OUTPUT
Prefix of the output file.
-h, --help show this help message and exit
--version show program's version number and exit
Example usage (xiFDR version < 2.2.1):
python xiFdrExporter_msannika.py XLpeplib_Beveridge_QEx-HFX_DSS_R1.xlsx
Or using the Windows binary:
xiFdrExporter_msannika.exe XLpeplib_Beveridge_QEx-HFX_DSS_R1.xlsx
Example usage (xiFDR version 2.2.1 or later):
python xiFdr2.2.1Exporter_msannika.py XLpeplib_Beveridge_QEx-HFX_DSS_R1.xlsx
Or using the Windows binary:
xiFdr2.2.1Exporter_msannika.exe XLpeplib_Beveridge_QEx-HFX_DSS_R1.xlsx
Export to PyXlinkViewer for pyMOL
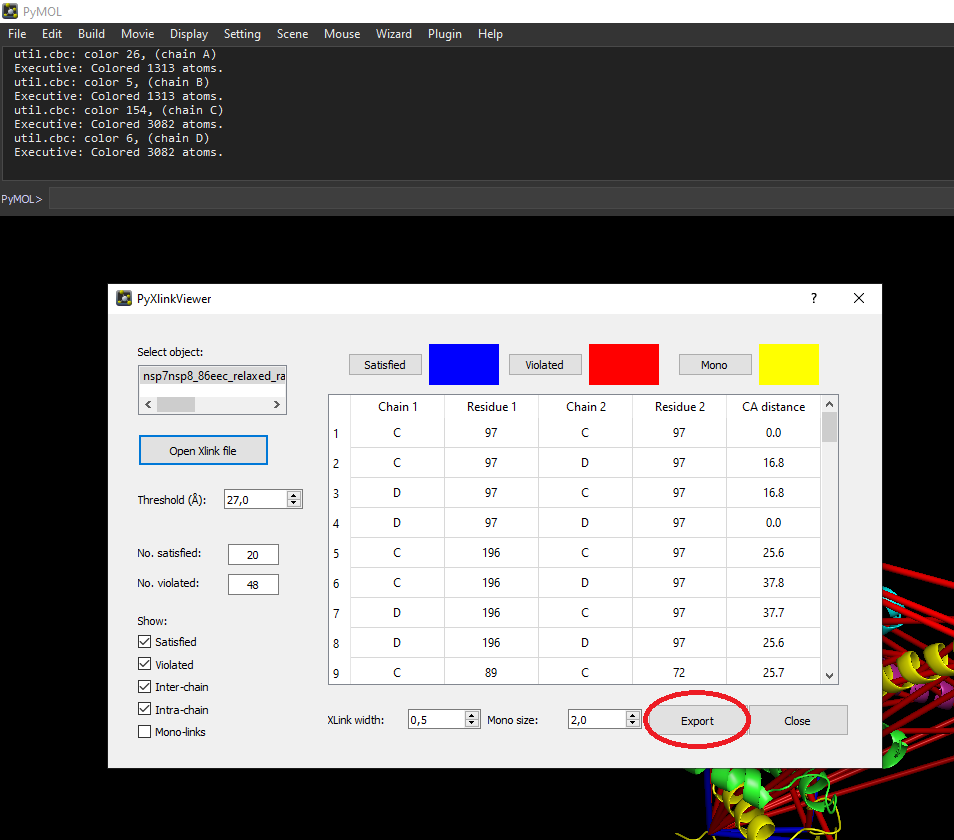
A schematic workflow of the implementation can be seen in this figure.
EXPORTER DESCRIPTION:
A script to export MS Annika results to PyXlinkViewer format for visualizing
crosslinks in pyMOL.
USAGE:
pyXlinkViewerExporter_msannika.py f [f ...]
[-pdb PDB_FILE]
[-go GAP_OPEN_PENALTY]
[-ge GAP_EXTENSION_PENALTY]
[-si SEQUENCE_IDENTITY]
[-allowxlmismatch]
[-o OUTPUT]
[-h]
[--version]
positional arguments:
f MS Annika crosslink result files in Microsoft Excel
format (.xlsx) to process.
required arguments:
-pdb PDB_FILE, --pdb PDB_FILE
PDB file of the structure that crosslinks should be
exported to/or 4-letter identifier from the PDB
(structure will be retrieved from the internet).
optional arguments:
-go GAP_OPEN_PENALTY, --gap_open GAP_OPEN_PENALTY
Gap open penalty for sequence alignment.
Default: -10
-ge GAP_EXTENSION_PENALTY, --gap_extension GAP_EXTENSION_PENALTY
Gap extension penalty for sequence alignment.
Default: -1
-si SEQUENCE_IDENTITY, --sequence_identity SEQUENCE_IDENTITY
Sequence identity threshold in percent to consider two
aligned sequences as matching.
Default: 80
-allowxlmismatch, --allowxlmismatch
Flag to report crosslinks that don't link to a crosslink
site in the PDB sequence.
Default: Do not report such crosslinks.
-ic, --ignore_chains
Ignore specific chains in the PDB file.
Default: No chains are ignored.
-h, --help show this help message and exit
-o OUTPUT, --output OUTPUT
Prefix of the output files.
--version show program's version number and exit
Example usage:
python pyXlinkViewerExporter_msannika.py "202001216_nsp8_trypsin_XL_REP1.xlsx" "202001216_nsp8_trypsin_XL_REP2.xlsx" "202001216_nsp8_trypsin_XL_REP3.xlsx" --pdb 6yhu.pdb -o test
Or using the Windows binary:
pyXlinkViewerExporter_msannika.exe "202001216_nsp8_trypsin_XL_REP1.xlsx" "202001216_nsp8_trypsin_XL_REP2.xlsx" "202001216_nsp8_trypsin_XL_REP3.xlsx" --pdb 6yhu.pdb -o test
Export to XLMS-Tools
XLMS-Tools uses the same input format as pyXlinkViewer. Please use the pyXlinkViewer exporter to export your results to XLMS-Tools format.
Export to XMAS for ChimeraX
Visualization of crosslinks with XMAS in ChimeraX works without the need of an additional exporter, the Microsoft Excel files exported from Proteome Discoverer can be used directly as evidence files within XMAS.
Export to PAE Viewer
Evaluating predicted structures (e.g. structures created with AlphaFold2) using cross-linking data can easily be done using PAE Viewer. Exporting MS Annika results to the input format of PAE Viewer requires first exporting to pyXlinkViewer (pyMOL) and then exporting crosslinks from pyXlinkViewer to CSV, as shown in the pyMOL screenshot below:
The exporter takes the following arguments:
EXPORTER DESCRIPTION:
A script to export MS Annika results from pyXlinkViewer to PAE Viewer input
files (CSV).
USAGE:
PAEViewerExporter_msannika.py f [f]
[-t DISTANCE]
[-o OUTPUT]
[-h]
[--version]
positional arguments:
f Crosslinks exported from pyXlinkViewer in csv format.
optional arguments:
-t DISTANCE, --threshold DISTANCE
threshold (float) that specifies if a crosslink
satisfies the crosslinker-specific distance constraint.
-o OUTPUT, --output OUTPUT
Prefix of the output file.
-h, --help show this help message and exit
--version show program's version number and exit
Example usage:
python PAEViewerExporter_msannika.py pyXlinkViewer_export.csv
Or using the Windows binary:
PAEViewerExporter_msannika.exe pyXlinkViewer_export.csv
Mapping crosslinks to 3D structures is often ambiguous as the same peptide may appear in more than one chain, creating several possible cross-linked residue pairs. If you only want to find the shortest residue pair/crosslink (e.g. for validating a 3D structure) please check out find_shortest.py in the Crosslink_Utils repo.
To use the xiNET and xiVIEW exporters in Proteome Discoverer, add a "Scripting Node" from the "Post-Processing" tab in the "Workflow Nodes" window to your consensus workflow. You need to specify the following parameters in the Scripting Node:
- Path to Executable: Path of the python installation e.g.
C:\Users\Username\AppData\Local\Programs\Python\Python37\python.exe - Command Line Arguments: Path of the exporter script from
scripting_nodesand%NODEARGS%e.g.C:\Users\Username\Documents\PDScriptingNodes\xiViewExporter_msannikaPD.py %NODEARGS% - Requested Tables and Columns: Copy and paste the contents of
pd_tables.txt
Re-running the consensus worklflow should create the xiNET/xiVIEW files in the study directory.
Tested with Proteome Discoverer 2.5 (version 2.5.0.400).
If you are using the exporters please cite:
MS Annika 2.0 Identifies Cross-Linked Peptides in MS2–MS3-Based Workflows at High Sensitivity and Specificity
Micha J. Birklbauer, Manuel Matzinger, Fränze Müller, Karl Mechtler, and Viktoria Dorfer
Journal of Proteome Research 2023 22 (9), 3009-3021
DOI: 10.1021/acs.jproteome.3c00325
If you are using MS Annika please cite:
MS Annika 2.0 Identifies Cross-Linked Peptides in MS2–MS3-Based Workflows at High Sensitivity and Specificity
Micha J. Birklbauer, Manuel Matzinger, Fränze Müller, Karl Mechtler, and Viktoria Dorfer
Journal of Proteome Research 2023 22 (9), 3009-3021
DOI: 10.1021/acs.jproteome.3c00325
or
MS Annika: A New Cross-Linking Search Engine
Georg J. Pirklbauer, Christian E. Stieger, Manuel Matzinger, Stephan Winkler, Karl Mechtler, and Viktoria Dorfer
Journal of Proteome Research 2021 20 (5), 2560-2569
DOI: 10.1021/acs.jproteome.0c01000