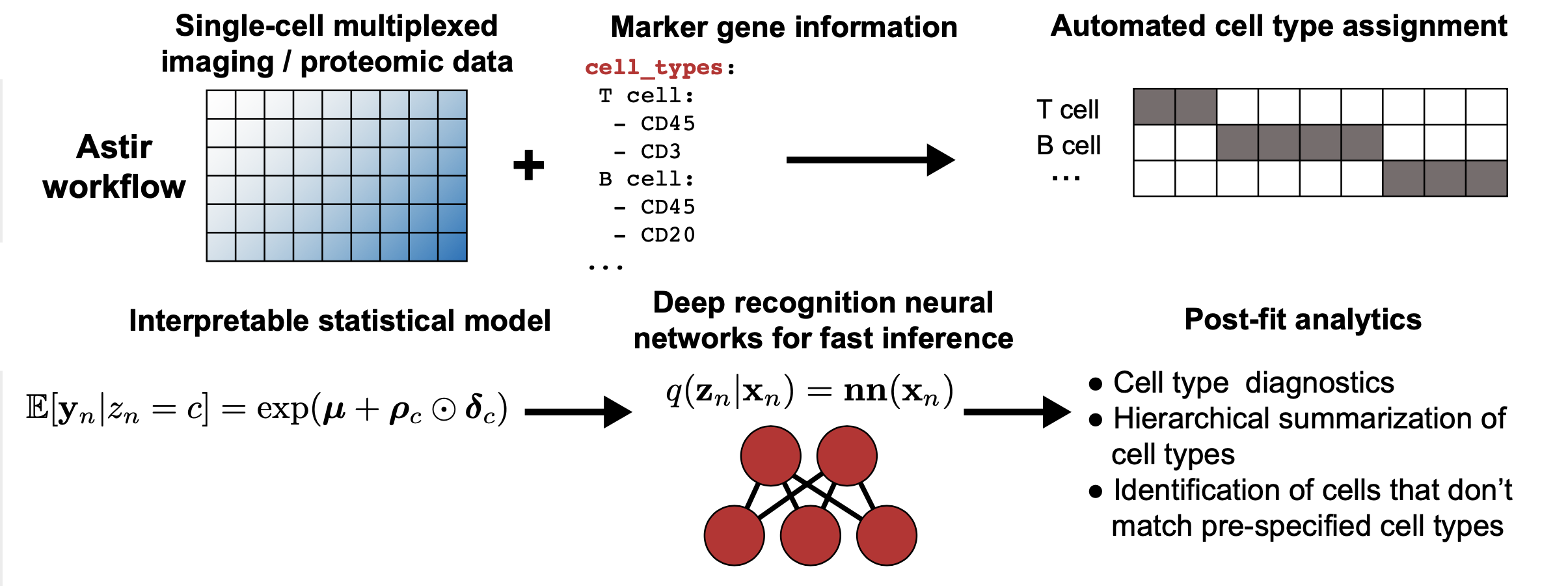
astir is a modelling framework for the assignment of cell type across a range of single-cell technologies such as Imaging Mass Cytometry (IMC). astir is built using pytorch and uses recognition networks for fast minibatch stochastic variational inference.
Key applications:
- Automated assignment of cell type and state from highly multiplexed imaging and proteomic data
- Diagnostic measures to check quality of resulting type and state inferences
- Ability to map new data to cell types and states trained on existing data using recognition neural networks
- A range of plotting and data loading utilities
Launch the interactive tutorial:
See the full documentation and check out the tutorials.
Jinyu Hou, Sunyun Lee, Michael Geuenich, Kieran Campbell
Lunenfeld-Tanenbaum Research Institute & University of Toronto
Lunenfeld-Tanenbaum Research Institute & University of Toronto