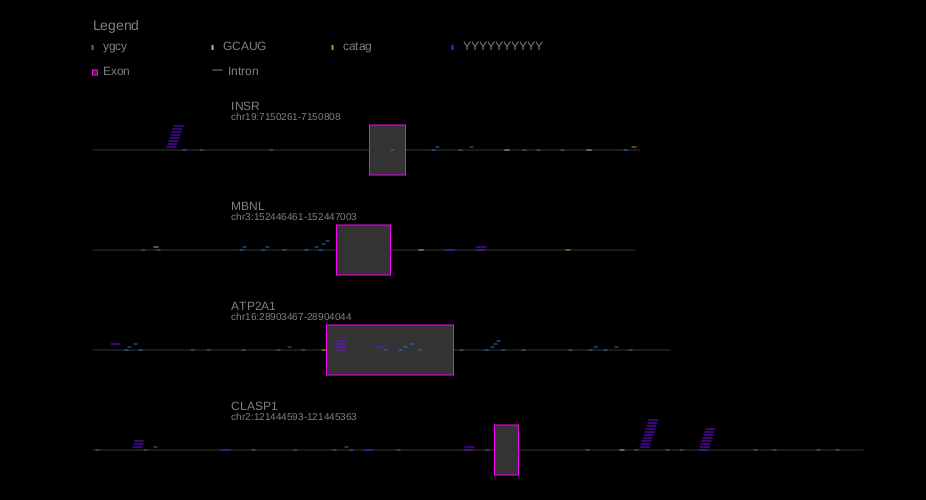
Motif mark generates an image of a gene, indicating locations of exons, introns, and motifs.
Motif mark takes two files as input: a fasta file and a motif file (with each motif on a single line).
git clone <repo>
The required dependencies for establishing a new conda environment are located in the environment.yml file. Replace $CONDA_ENV_NAME with your desired name for the motif-mark conda environemnt.
conda create --name $CONDA_ENV_NAME -f environment.yml
conda activate $CONDA_ENV_NAME
Run the following command to generate your image, replacing the variables with the corresponding values:
./motif-mark-oop.py -f $FASTA_FILE -m $MOTIF_FILE
By default, the python script will place the image in a directory output/ adjacent to the directory where the $FASTA_FILE is located. The default name of the image file will be the same as the $FASTA_FILE file, but with .png extension.
If you wish to use an alternative output directory, specify the location using the -d flag. If you wish for the file to have an alternative name, specify the name using the -o flag.