Rails app which exposes a RESTful API for a Long Read LIMS
- Add details aboout traction ui
- importing samples from external sources currently:
- Sequencescape
- Samples extraction
- creating libraries
- creating pools of libraries with support for multiplexing
- creating sequencing runs
- printing sample sheets
- supports the following technologies:
- Saphyr
- Pacbio
- ONT (In progress)
- Ruby (check
.ruby-versionfor the version) and ruby version manager - Bundler
gem install bundler - Graphviz (for mac OS
brew install graphviz) - OpenSSL
- MySQL
brew install mysql@8.0
- Run
bin/setup
This will:
- Run bundle install
- Create copies of any .example files
- Create the database
- Seed basic data such as library types and tags.
Note:
-
If the mysql-client cannot be found (possibly due to the homebrew installation path not being the default), the
mysql2gem will fail to install with the exception:... /Users/<user>/.rbenv/versions/3.1.2/lib/ruby/3.1.0/mkmf.rb:1086:in `block in find_library': undefined method `split' for nil:NilClass (NoMethodError) ... -
The fix is to add mysql@8.0 binaries to the PATH [^1] similar to the following:
export PATH=$PATH:(brew --prefix)/opt/mysql@8.0/bin
The database should have been generated as part of the installation step above.
If you need to create the database afresh: bundle exec rails db:reset.
The following tasks are run automatically by rails db:seed at the end of the
setup process
- To create all the supported library types:
bundle exec rails library_types:create - To create all the supported data types:
bundle exec rails data_types:create - To create a set of enzymes (needed for saphyr dummy runs):
bundle exec rails enzymes:create - To create all of the pacbio tags (needed for pacbio dummy runs):
bundle exec rails tags:create:pacbio_all - To create pacbio Qc Assay Types:
bundle exec rails qc_assay_types:create - To create pacbio smrt link versions:
bundle exec rails smrt_link_versions:create - To create ont instruments:
bundle exec rails ont_instruments:create - To create the tree of life tubes report view:
bundle exec rails tol_tubes_report_view:create
To create pacbio dummy runs: bundle exec rails pacbio_data:create
To create saphyr dummy runs: bundle exec rails saphyr_runs:create
To create ont dummy runs: bundle exec rails ont_data:create
To create dummy printers: bundle exec rails dummy_printers:create
To drop the database bundle exec rails db:drop
To run the unit tests run rspec. bundle exec rspec
We use rubocop to keep the code clean bundle exec rubocop
To run the rails application bundle exec rails s
When running with Traction-UI, UI expects the service to be on port 3100. PORT=3100 rails s
Sending messages is disabled by default but if you would like to test messages, install a broker
(RabbitMQ) and update the config in config/bunny.yml by enabling the development settings.
After installing RabbitMQ, you will need to create the exchange you will be sending messages over. You can do this by issuing a command in your terminal such as
rabbitmqadmin declare exchange name="bunny.examples.exchange" type="topic"making sure you match the exchange name with the one specified in config/bunny.yml.
A web interface to administrate RabbitMQ is always available at http://localhost:15672/ once the service is running.
Sample sheets can be generated for upload to SMRT Link.
For more details see the notes on the Sample Sheet Generator.
You may wish to enable the provided git commit hook if you want to be notified of rubocop issues in the files you've edited before they are committed.
To do this, refer to the documentation in .githooks/README.txt.
To see all the commands available from rails: bundle exec rails -T
Sample sheets can be downloaded en masse for debugging and development purposes using the download_sample_sheets command in the scripts directory. See the scripts README for more information.
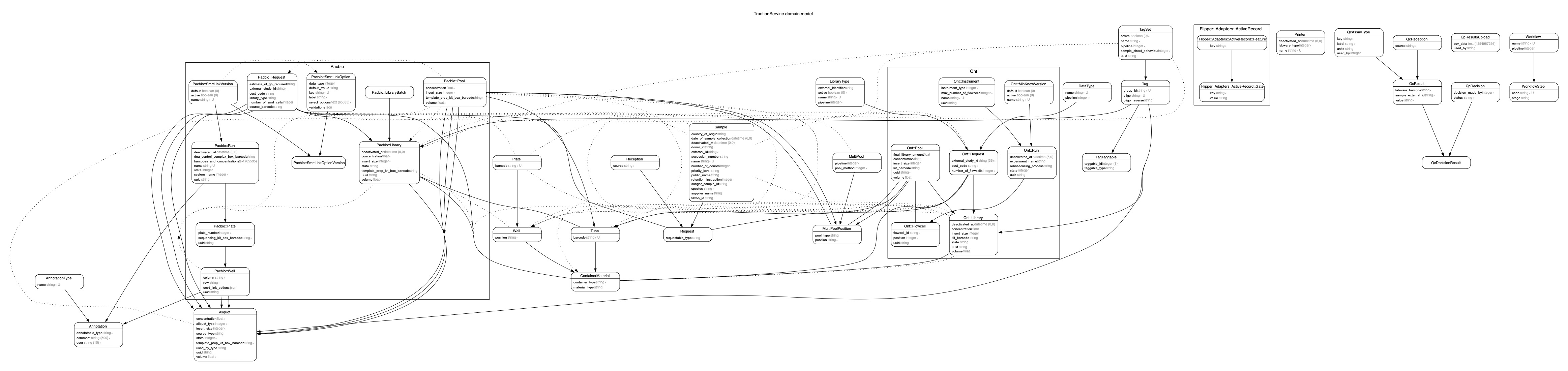
An ERD was created using the rails-erd gem by executing: bundle exec erd
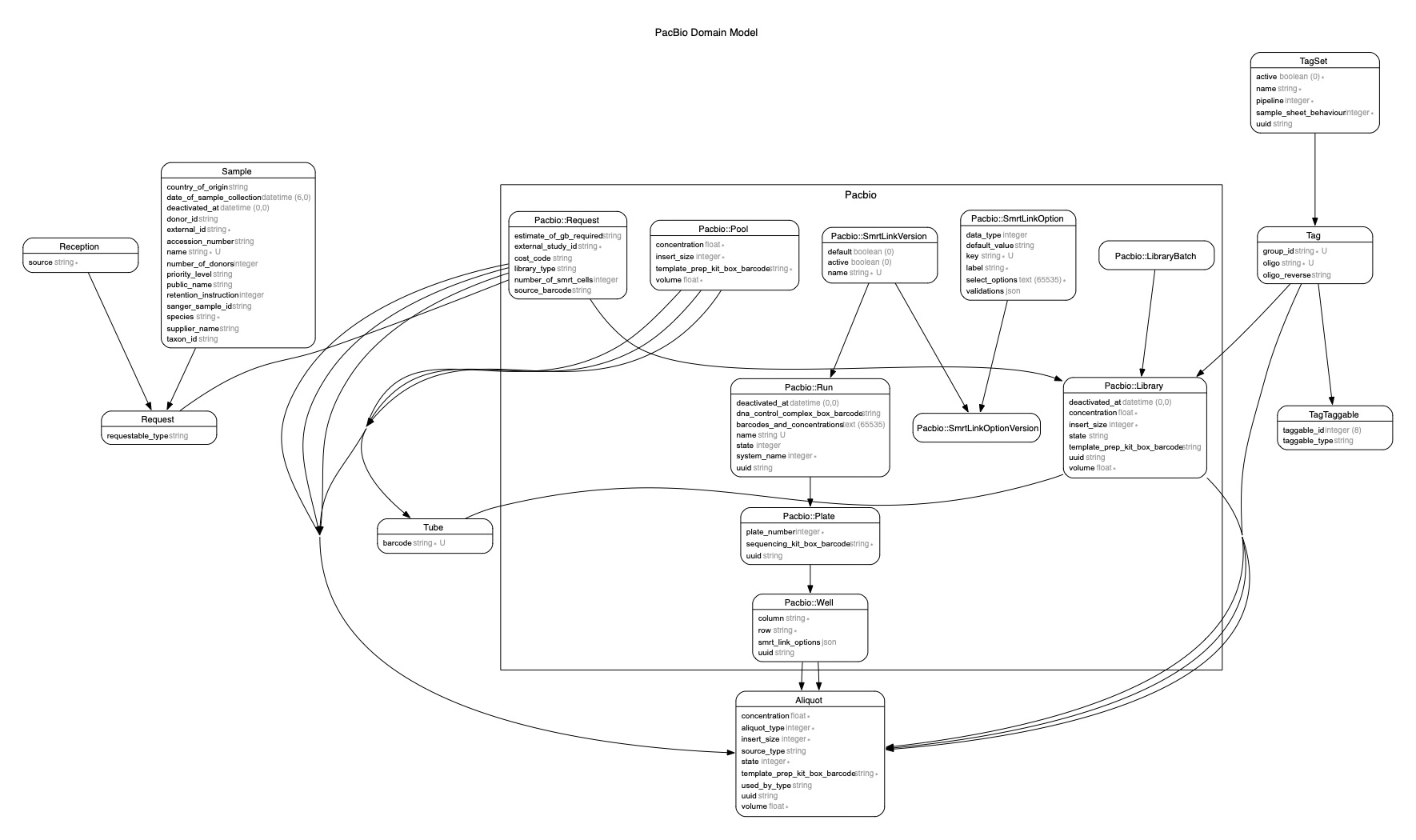
A custom PacBio ERD was created using the .erd.pacbio.erdconfig config by executing: bundle exec erd --config .erd.pacbio.erdconfig
Should the ERD fail to generate with the message Failed: ArgumentError: bad value for range, apply the patch below:
diff --git a/app/models/ont/run.rb b/app/models/ont/run.rb
index 4dd7083e..d3bde752 100644
--- a/app/models/ont/run.rb
+++ b/app/models/ont/run.rb
@@ -42,7 +42,7 @@ module Ont
message: :run_min_flowcells
}
validates :flowcells, length: {
- maximum: :max_number_of_flowcells, if: :max_number_of_flowcells,
+ maximum: 2, # :max_number_of_flowcells, if: :max_number_of_flowcells, # TODO: fix failing ERD
message: :run_max_flowcells
}On merging a pull request into develop, a release will be created with the tag/name <branch>/<timestamp>
Update .release-version with major/minor/patch. On merging a pull request into master, a release will be created with the release version as the tag/name
See Confluence for further information
We've used Yard as the documentation tool to document the service. This can be accessed via https://sanger.github.io/traction-service/.
To query all the @todo items in Yard, the following command can be used:
bundle exec yard list --query '@todo'It is possible to add comprehensive documentation about the areas in each application. For example, in this repository, we have added a documentation on how volume tracking works. The process to add these sub-documentation is as follows.
Prerequisites
Install a recent version of Python. Python is used for the mkdocs tool we use to generate documentation. Please install mkdocs using pip install mkdocs following installing Python.
Install the following dependencies:
pip install mkdocs-material
pip install mkdocs-glightbox
pip install mkdocs-git-revision-date-localized-plugin- Navigate to
documentationdirectory. - Create a new
mkdocsdocumentation withmkdocs new <sub documentation name>. Please be aware that thissub documentation namewill be the route you'll have use to navigate to the documentation. For example, if it's volume-tracking, you'll have to navigate tosanger.github.io/traction-service/volume-trackingto access documentation. - Use markdown to write up the documentation in the newly generated directory's
docsubdirectory.
A CI action will automatically push the documentation upon master releases. If you want to deploy the documentation manually, you can the manual dispatch action.