HistoSeg : Quick attention with multi-loss function for multi-structure segmentation in digital histology images
Paper was presented at 12th International Conference on Pattern Recognition Systems (ICPRS), 2022 École Nationale Supérieure des Mines de Saint-Étienne, France
DOI: 10.1109/ICPRS54038.2022.9854067
Copyrights has been given to IEEE. IEEE Xplore link is https://ieeexplore.ieee.org/document/9854067
Please Cite it as following
@inproceedings{wazir2022histoseg,
title={HistoSeg: Quick attention with multi-loss function for multi-structure segmentation in digital histology images},
author={Wazir, Saad and Fraz, Muhammad Moazam},
booktitle={2022 12th International Conference on Pattern Recognition Systems (ICPRS)},
pages={1--7},
year={2022},
organization={IEEE}
}
This repo contains the code to Test and Train the HistoSeg
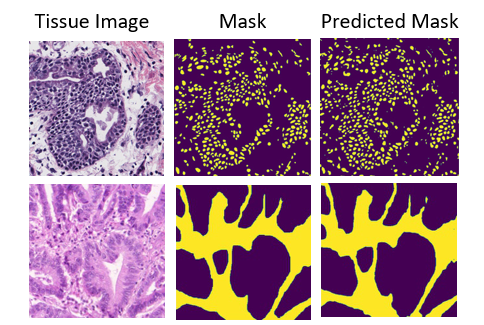
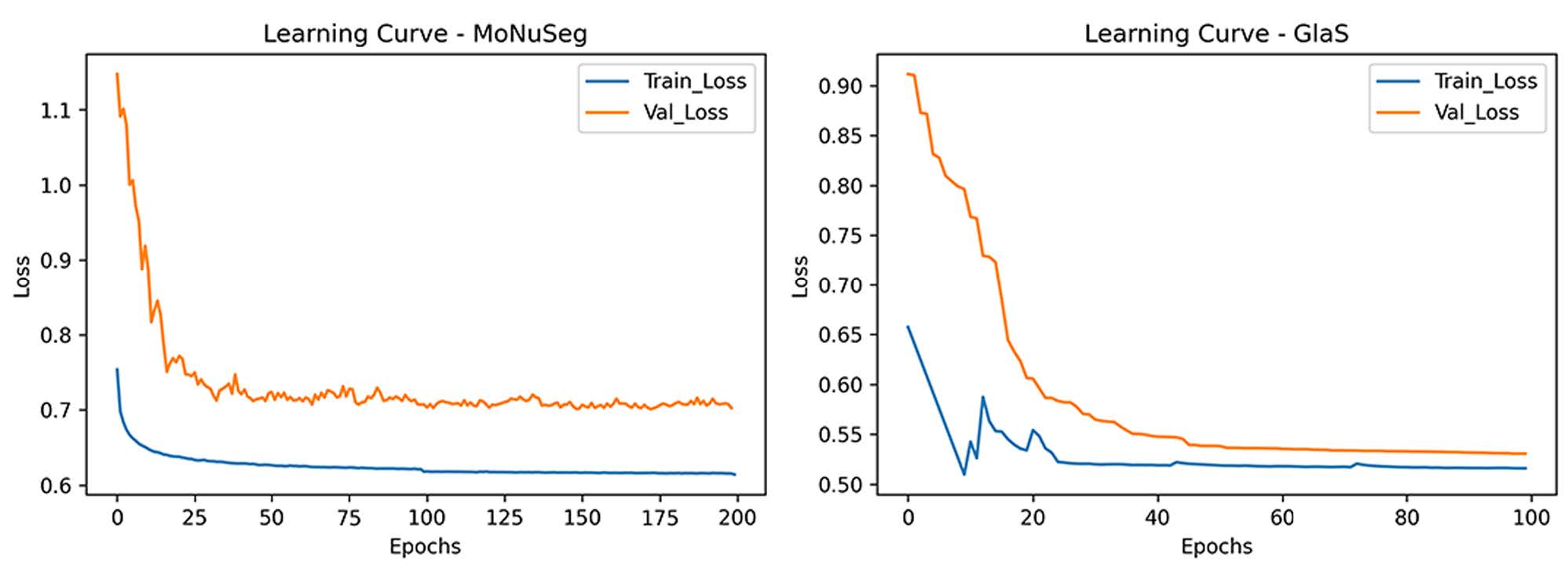
HistoSeg is an Encoder-Decoder DCNN which utilizes the novel Quick Attention Modules and Multi Loss function to generate segmentation masks from histopathological images with greater accuracy.
| MoNuSeg | GlaS | ||||
|---|---|---|---|---|---|
| F1 | IoU | Dice | F1 | IoU | Dice |
| 75.08 | 71.06 | 95.20 | 98.07 | 76.73 | 99.09 |
link: https://monuseg.grand-challenge.org/
link: https://warwick.ac.uk/fac/cross_fac/tia/data/glascontest/
For MoNuSeg Dataset link: https://github.com/saadwazir/HistoSeg/blob/main/HistoSeg_MoNuSeg_.h5
For GlaS Dataset link: https://github.com/saadwazir/HistoSeg/blob/main/HistoSeg_GlaS_.h5
After downloading the dataset you must generate patches of images and their corresponding masks (Ground Truth), & convert it into numpy arrays or you can use dataloaders directly inside the code. Note: The last channel of masks must have black and white (0,1) values not greyscale(0 to 255) values. you can generate patches using Image_Patchyfy. Link : https://github.com/saadwazir/Image_Patchyfy
For example to train HistoSeg on MoNuSeg Dataset, the distribution of dataset after creating pathes
X_train 1470x256x256x3
y_train 1470x256x256x1
X_val 686x256x256x3
y_Val 686x256x256x1
You just need to resize the images and their corresponding masks (Ground Truth) into same size i.e all the samples must have same resolution, and then convert it into numpy arrays.
For example to test HistoSeg on MoNuSeg Dataset, the shapes of dataset after creating numpy arrays are
X_test 14x1000x1000x3
y_test 14x1000x1000x1
pip install matplotlib
pip install seaborn
pip install tqdm
pip install scikit-learn
pip install scikit-image
conda install -c conda-forge tensorflow==2.7
pip install keras==2.2.4
To train HistoSeg use the following command
python HistoSeg_Train.py --train_images 'path' --train_masks 'path' --val_images 'path' --val_masks 'path' --width 256 --height 256 --epochs 100 --batch 16
To test HistoSeg use the following command
python HistoSeg_Test.py --images 'path' --masks 'path' --weights 'path' --width 1000 --height 1000
For example to test HistoSeg on MoNuSeg Dataset with trained weights, use the following command
python HistoSeg_Test.py --images 'X_test_MoNuSeg_14x1000x1000.npy' --masks 'y_test_MoNuSeg_14x1000x1000.npy' --weights 'HistoSeg_MoNuSeg_.h5' --width 1000 --height 1000