This is a standalone package, however it requires python3 and java to run.
python3 --version
Should return something like:
Python 3.x.y
(otherwise you will need to install it)
java --version
Should return something like:
openjdk 11.0.18 2023-01-17
OpenJDK Runtime Environment (build 11.0.18+10-post-Ubuntu-0ubuntu122.04)
OpenJDK 64-Bit Server VM (build 11.0.18+10-post-Ubuntu-0ubuntu122.04, mixed mode, sharing)
(otherwise you will need to install it)
Once you have python3 and java on your machine, clone the repository or download it on your computer.
Change directory to the folder where you downloaded the zip:
cd {folder}
Unzip file and enter folder:
unzip LigandMapper.py-main.zip
cd LigandMapper.py-main
And run:
python3 installMe.py
During the installation you will be prompted to enter the password for sudo because the file needs to create a link in the /usr/bin/ folder.
After a successful installation you can remove the zip and the LigandMapper.py-main.zip directory.
In case the installation fails:
-
You can run the program from the folder itself because it's already an executable with a python3 shebang.
-
And if you want it accessible to the command line as a command, then:
- choose a directory from your $PATH (echo $PATH) - ex. /usr/bin/
- create a symbolic link there (sudo ln -s ./LigandMapper/LigandMapper.py /usr/bin/)
- then add add permissions to the whole package (sudo chmod -R 777 ./)
- do not move or delete the folder!
dir_path=$(find ~ -name 'LigandMapper' -type d) fileLM=$dir_path"/LigandMapper.py" sudo ln -s $fileLM /usr/bin/ sudo chmod -R 777 ./
Note: The ouput is to the standard error.
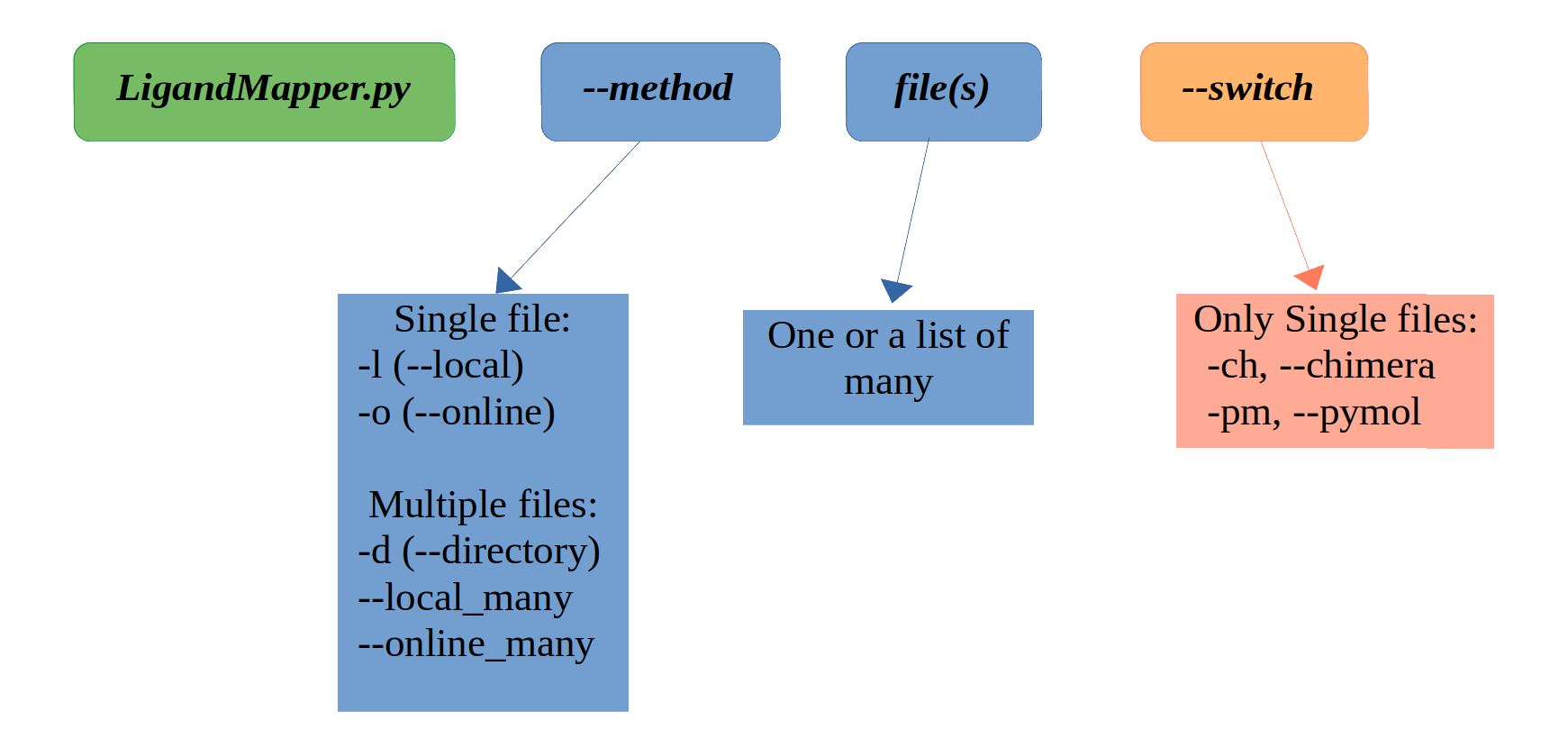
usage: LigandMapper.py [-h] [-l {file}.pdb] [-o {pdb_name}] [-d {direcory_name}] [--local_many [{file1}.pdb {file2}.pdb ...]] [--online_many [{pdb_name1} {pdb_name2} ...]] [-v] [-ch]
[-pm]
LigandMapper.py is a python script build to predict ligand binding sites of proteins from their .pdb files.
optional arguments:
-h, --help show this help message and exit
-l {file}.pdb, --local {file}.pdb
One pdb local file.
-o {pdb_name}, --online {pdb_name}
Get a pdb file from the pdb server and analyse that.
-d {direcory_name}, --directory {direcory_name}
Analyse all files located in one local directory.
--local_many [{file1}.pdb {file2}.pdb ...]
Analyse many local pdb files.
--online_many [{pdb_name1} {pdb_name2} ...]
Get many pdb files from the pdb server and analyse them.
-v, --verbose Get more detailed output of the process to the standard error.
-ch, --chimera Open chimera immediately when file is ready to be visualised. Only applies to local and online SINGLE file.
-pm, --pymol Open pymol immediately when file is ready to be visualised. Only applies to local and online SINGLE file.
Tutorials can be found here.
- If you have one protein structure pdb file (ex. 1gln.pdb) on your computer and you want to predict the ligand binding pockets, run the --local (-l) method:
LigandMapper.py -l 1gln.pdb
- If you know whats the pdb name but you don't have the pdb file downloaded, you can automatically download it and run the analysis with the --online (-o) method:
LigandMapper.py -o 1gln
- If you have a directory of pdb files you want to analyse, run the --directory (-d) method:
LigandMapper.py -d directory_name/
- Or, my advice is, to run this once you have changed into the directory with the files (that way all the output will be stored in the same directory):
cd directory_name/
LigandMapper.py -d ./
- If you have many local files that you want to analyse, but they're not exclusively gropued in one directory, you can just list them with the --local_many method:
LigandMapper.py --local_many 1gln.pdb 2ew2.pdb subfol1/1gln.pdb
- If you want to download from the pdb server and analyse many pdb files, list them with the --online_many method:
LigandMapper.py --online_many 1gn3 2ew2 1gln
- The output can directly visualised with the --chimera (-ch) and --pymol (-pm) switch, given that you have them installed on your computer, by including the switch when running the comand. Note: this only woks for the single-file methods (-l (--local) and -o (--online)); for the --directory, --online_many and --local_many you need to open the visualisation cmd files manually (see: Output below).
Ex.
LigandMapper.py -l 1gln.pdb -ch
LigandMapper.py -o 1gln -pm
A prediction for a file {pdb}.pdb will create the following structure in the folder in which LigandMapper.py was executed.
predict_{pdb}/
├── {pdb}.pdb_predictions.tsv
└── visualizations/
├── chimera_{pdb}.cmd
├── {pdb}.pdb.pml
└── data/
├── {pdb}.pdb_points.pdb.gz
└── {pdb}.pdb
The tsv file lists the predicted pockets in order of their score (probability, see theoretical background ). Each pocket has the following attributes:
-
rank
-
score
-
probability
-
sas_points - (int) number of solvent accessible surface points
-
surf_atoms - (int) integer of the number of surface atoms
-
center_x - (float) the predicted pockets x center
-
center_y - (float) the predicted pockets y center
-
center_z - (float) the predicted pockets z center
-
residue_ids - (py dict) the residue sequence numbers that create the pocket { Chain : [ residue sequence numbers ] }
-
residue_names - (py dict) the residue names that create the pocket { Chain : [ residue names ] }
-
residue_types - (py dict) the character of the residues that create the pocket { Chain : [ characters ] }
- 'N' represents non-polar amino acids
- 'P' represents polar amino acids
- '+' represents positively charged amino acids
- '-' represents negatively charged amino acids
- '0' a specific residue for which there is no info in the program
-
surf_atom_ids - (py list) the atom serial number of all the atoms that are on the surface of the pocket
Information from the PDB is taken in this fashion:
| Columns | Data | Justification | Data Type |
|---|---|---|---|
| 1-4 | "ATOM" | left | character |
| 7-11 | Atom serial number | right | integer |
| 13-16 | Atom name | left* | character |
| 17 | Alternate location indicator | - | character |
| 18-20 | Residue name | right | character |
| 22 | Chain identifier | - | character |
| 23-26 | Residue sequence number | right | integer |
| 27 | Code for insertions of residues | - | character |
| 31-38 | X orthogonal Angstrom coordinate | right | floating |
| 39-46 | Y orthogonal Angstrom coordinate | right | floating |
| 47-54 | Z orthogonal Angstrom coordinate | right | floating |
| 55-60 | Occupancy | right | floating |
| 61-66 | Temperature factor | right | floating |
| 73-76 | Segment identifier (optional) | left | character |
| 77-78 | Element symbol | right | character |
| 79-80 | Charge (optional) | - | character |
Has neccessary information to create the visualisations in chimera. The pockets are saved as selections titled "Pocket{NUM}" and colored untill the 18th pockets.
The colors are ranked the same in all output, so it can be a visual aid for quick understanding of the pockets' rankings. This is true only for the Chimera file, not for the PyMol, because in PyMol the pockets and their colors can be easily viewed in the side panel.
| Color | Rank |
|---|---|
| red | 1 |
| orange | 2 |
| yellow | 3 |
| green | 4 |
| cyan | 5 |
| blue | 6 |
| medium blue | 7 |
| purple | 8 |
| hot pink | 9 |
| magenta | 10 |
| white | 11 |
| gray | 12 |
| black | 13 |
| tan | 14 |
| slate gray | 15 |
| dark khaki | 16 |
| plum | 17 |
| rosy brown | 18 |
run :
chimera {path}/predict_{pdb}/visualizations/chimera_{pdb}.cmd
In PyMol the pockets and their colors are conveniently displayed in the side panel. run:
pymol {path}/predict_{pdb}/visualizations/{pdb}.pdb.pml
The theorethical background can be found here.
Analysis of the perfomance can be found here.
This software is a lightweight version of p2rank.
- Krivak R, Hoksza D. P2Rank: machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. Journal of Cheminformatics. 2018 Aug.
- Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS
- Connolly M. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221(4612):709–13.
- Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006 Sep 24;6:19.
- Krivák R, Hoksza D. Improving protein-ligand binding site prediction accuracy by classification of inner pocket points using local features. J Cheminform. 2015 Apr 1;7:12.
- Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009 Jun 2;10:168.
- RICHARDS, E M. (1977). Ann. Rev. Biophys. Bioeng. 6, 151-176.
- Zheng X, Gan L, Wang E, Wang J. Pocket-based drug design: exploring pocket space. AAPS J. 2013 Jan;15(1):228-41.