-
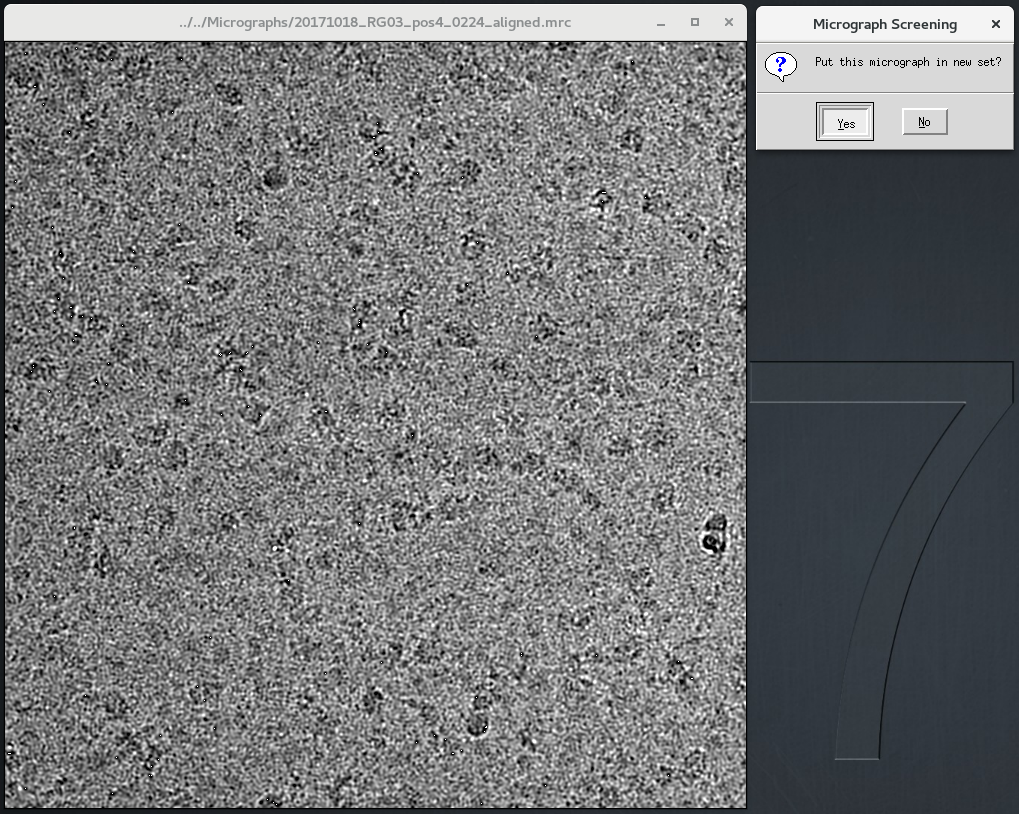
OJcryo_relion_screen: A simple interactive micrograph screening program that allows the removal of dirty/unwanted images from a cryo-EM image pool.
-
OJcryo_relion_rm_duplicates: Removes any duplicate particles from a Relion join_particles.star file.
-
OJcryo_m2_tiff_batch: Simple implementation of a TIFF batch mode for motioncor2 (requires GPU).
-
OJcryo_unblur_tiff: Implements a TIFF batch and parallel-processing functionality for unblur; adjusts using gain reference, bins, and then unblurs (tif2mrc->clip->binvol->unblur).
OJcryo is a project I have been slowly adding to over the past two years as I have written helper scripts for my work in processing cryo-em data. I hope you find it useful as some of these have saved me LOTS of manual effort. Enjoy! -np
DEPENDENCIES: RELION
- Navigate to an Import/ folder in your RELION project that has a micrographs.star inside.
- Run: OJcryo_relion_screen.py <output_filename.star (optional)>
- One-by-one, each micrograph in this folder's micrographs.star will be displayed. Once you have made a decision on whether to keep it or not, click the window "X" (or press Escape).
- A new window will pop up. To save the micrograph in your set, select "Yes" (or press Enter). Otherwise select "No" (or press Escape).
NOTE: OJcryo_relion_screen will run best when loading from a mounted drive, preferably an SSD. Screening over ssh will leave you pulling your hair out!
- Navigate to a JoinStar/ folder that contains a join_particles.star or join_mics.star file.
- RUN...
- (for particles): OJcryo_relion_rm_duplicates -p <starfile>
- (for micrographs): OJcryo_relion_rm_duplicates -m <starfile>
- Output:
- (for particles): join_particles_no_duplicates.star
- (for micrographs): join_mics_no_duplicates.star
Included example: Take a look at dupe_tester.star in the rm_duplicates folder. There are multiple entries for particle #314 (column 6). Run: OJcryo_relion_rm_duplicates -p dupe_tester.star Now look at the join_particles_no_duplicates.star -> NO DUPLICATES!
DEPENDENCIES: MOTIONCOR2 We capture images from the microscope as .tif files. Unfortunately, UCSF's MotionCor2 only corrects .mrc files in batch mode. This script jerry-rigs .tif batch mode retaining many of the optional arguments of MotionCor2.
There are many arguments possible for OJcryo_m2_tiff_batch so I simply encourage you to run the program with the -h tag to get the full usage. If you are unfamiliar with the correlating options terminology in MotionCor2 I would encourage you to read over their manual: http://msg.ucsf.edu/MotionCor2/MotionCor2-UserManual-05-03-2018.pdf
DEPENDENCIES: tif2mrc, clip, binvol, and unblur (v 1.0.2) Like Motioncor2, Niko Grigorieff's UNBLUR only does batch micrograph correction for .mrc files. With this program, not only is .tif batch mode implemented, but it also leverages subprocesses to align your movies up to 6x faster (!) (based on my prelim testing).
To run:
- Navigate to the directory where all your TIFFs are.
- Fill in the following command with your parameters:
- OJcryo_unblur_tiff <gainref> <cores> <ImgsPerStack> <A/pix> <DoseFilter?> <ExpsrPerFrm> <kV> <Pre-ExpsrAmount> <SaveAlignedFrames?> <ExpertOptions?>
- NOTE: My tests showed that the speed gained from more <cores> greatly diminishes after 16, but YMMV
- The aligned .mrc files will be written to a folder called "driftcorr_mrc\"