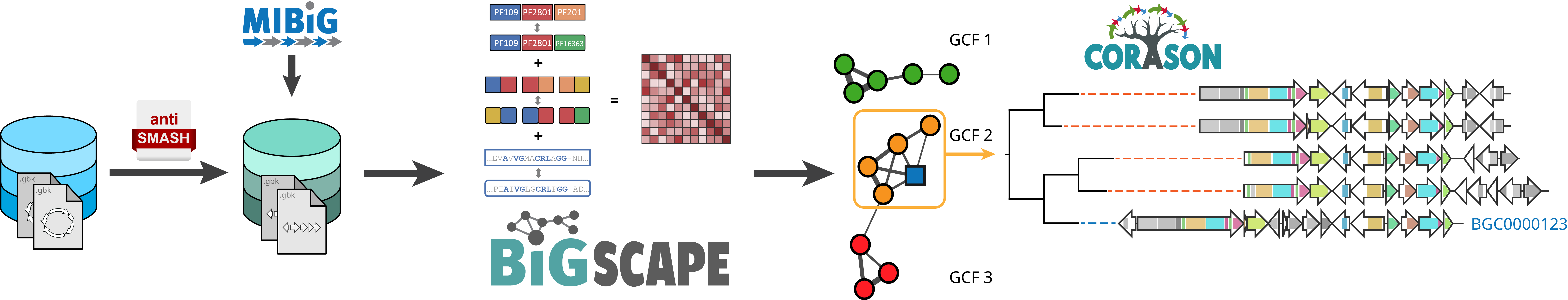
BiG-SCAPE (Biosynthetic Gene Similarity Clustering and Prospecting Engine) is a software package, written in Python, that constructs sequence similarity networks of Biosynthetic Gene Clusters (BGCs) and groups them into Gene Cluster Families (GCFs). BiG-SCAPE does this by rapidly calculating a distance matrix between gene clusters based on a comparison of their protein domain content, order, copy number and sequence identity.
As input, BiG-SCAPE takes GenBank files from the output of antiSMASH with BGC predictions, as well as reference BGCs from the MIBiG repository. As output, BiG-SCAPE generates tab-delimited output files, as well as a rich HTML visualization that includes multi-locus phylogenies of each Gene Cluster Family made using CORASON.
In principle, BiG-SCAPE can also be used on any other gene clusters, such as pathogenicity islands, secretion system-encoding gene clusters, or even whole viral genomes.
Learn more about BiG-SCAPE in the wiki and in the combined BiG-SCAPE/CORASON website.
If you find BiG-SCAPE useful, please cite us.