Visualization of small RNAseq data
cite:
Pantano Rubino L, Chapman B, Kirchner R et al. Characterization of the small RNA transcriptome using the bcbio-nextgen python framework [version 1; not peer reviewed]. F1000Research 2016, 5(ISCB Comm J):1627 (slides) (doi: 10.7490/f1000research.1112526.1)
Pantano L, Pantano Rubino F, Marti E and Ho Sui S. Visualization of the small RNA transcriptome using seqclusterViz [version 1; not peer reviewed]. F1000Research 2017, 6(ISCB Comm J):673 (poster) doi: 10.7490/f1000research.1114055.1
How to:
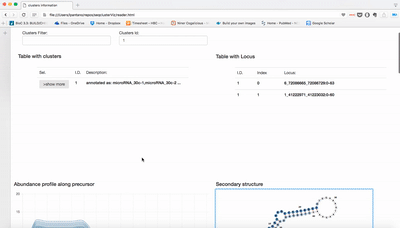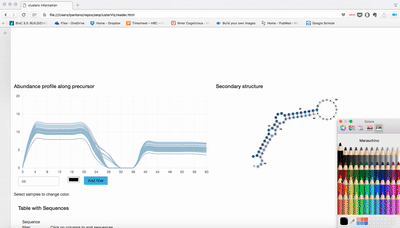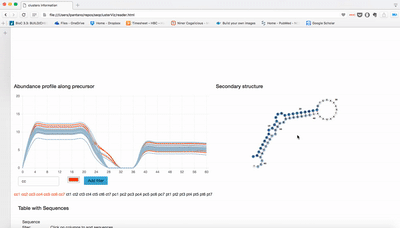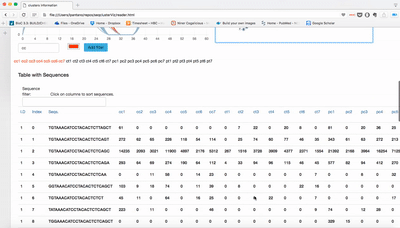
Visualization of seqcluster output for small RNA data. You can run seqcluster or bcbio-nextgen to get the output. With bcbio-nextgen the full analysis is much easier and you get plenty of other outputs. See an example of bcbio output here. This is the re-analysis of small RNA from patients with Parkinson's disease published in bioinformatics. Use the seqcluster.db to visualize this data with the tool.
Download this repo, unzip it, open index.html and use the tiny data data/example.db to test it!
See video: https://youtu.be/Zjzte8n2-Sg