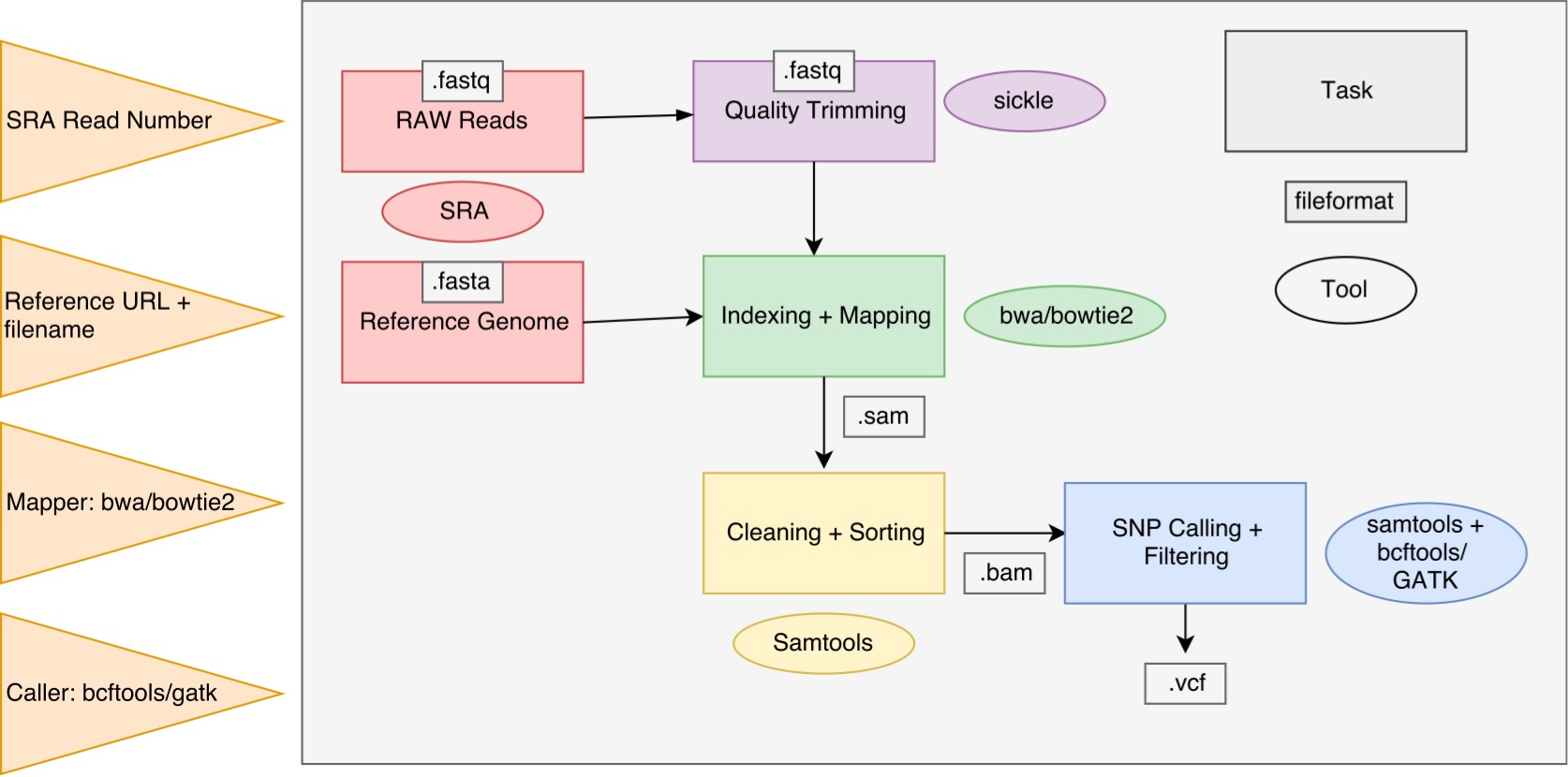
- Workflow "Language": GNU Make - Version 4.1
- Data accession: SRAToolkit - Version 2.8.2
- Quality Trimming: Sickle - Version 1.33
- Mapping:
- SNV Calling and preprocessing
As well as wget and gunzip. The workflow was tested on a Ubuntu 16.04 environment.
The workflow itself is based on GNU Make and can be easily called by:
make -f Makefile MAPPER="name_of_the_mapper" CALLER="name_of_the_caller" SRA="sra_number" REF_URL="ref_genome_ftp_url" REF_GENOME_FILE="unzipped_ref_genome_file_name"
The user can choose between two mappers, bwa and bowtie2, as well as between two variant calling tools, samtools/bcftools and gatk. In the following example call you use bwa to do the mapping and gatk to do the variant calling. Therefore we take the e-coli genome as the reference genome and the raw reads contained in the SRA number SRR3736639:
make -f Makefile MAPPER="bwa" CALLER="bcftools" SRA="SRR3736639" REF_URL="ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/845/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna.gz" REF_GENOME_FILE="GCF_000005845.2_ASM584v2_genomic.fna"
Attention:
- The SRA samples should contain both forward and reverse strands!
- If files are already existing in the folders the workflow generates, they wont be computed again. This allows an easy continuation of the computations
- BUT if you cancel the workflow right in between you should delete the corrupt files by hand!
In this case an attached bash-script (run_workflow.sh) can be easily adjusted to perform the workflow on multiple SRA samples with different map and variant call tools. This is an example of how the adjustment could look like:
ref_url="ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/845/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna.gz"
ref_file="GCF_000005845.2_ASM584v2_genomic.fna"
declare -a mappers=("bwa"
"bowtie2"
)
declare -a callers=("bcftools"
"gatk"
)
declare -a sras=("SRR3736622" "SRR3736623")
and finally run the bash script in the same folder as the Makefile of the workflow:
./run_workflow.sh
This would run the workflow for the two given samples four times: once with bwa und bcftools, once with bowtie2 and bcftools, once with bwa and gatk and last but not least with bowtie2 and gatk.
The workflow delivers a .vcf file for the corresponding SRA sample and its reference genome. If you use gatk for the variant calling the resulting .vfc file only contains the snps! The .vfc file generated by bcftools contains everything.