Python for RAW Sentinel2 data (PyRawS) is a powerful open-source Python package that provides a comprehensive set of tools for working with Sentinel-2 Raw data🔬.
1
It provides utilities for coarse spatial bands coregistration, geo-referencing, data visualization📊, and image processing🖼️.
The software is demonstrated on the first Sentinel-2 🛰️ Raw database for warm temperature hotspots 🔥 detection/classification, making it an ideal tool for a wide range of applications in remote sensing and earth observation🌍.
The package is written in Python and is open source💻, making it easy to use and modify for your specific needs.
The systme is based on pytorch, which be installed with CUDA support, to enable GPU acceleation.
NB: What we call raw data in this project are Sentinel-2 data generated by the decompression and metadata addition of Sentinel-2 L0 data. Because of that, with the exception of the effects due to onboard equalization and lossy compression, they are the most similar version of the rawest form of data acquired by the satellite's sensors. Both the compression and equalization are applied onboard the satellite to reduce the amount of data transmitted to the ground station. For easy naming convention, this repo refer to the term "Raw" as the products decompressed with ancillary information appended. For further information browse our paper at https://arxiv.org/abs/2305.11891 ↩
(Disclaimer: This project is currently under development.)
A demo showcasing PyRawS capabilities is available on the YouTube channel of Robin Cole
Table of Contents
- About the Project
- Content of the repository
- Installation
- PyRawS databases
- Raw events and Raw granules
- Quickstart
- Open a Raw event from PATH.
- Open a Raw event from database.
- Show Raw granules information of a Raw event
- Access a Raw_granule pixels
- Get a single Raw_granule from a Raw_event
- Superimpose Raw_granule bands
- How to perform the coarse coregisteration of Raw_granules
- How to get the coordinates of a Raw granule band
- Raw_event: database metadata
- Export a Raw_granule to TIF
- Glossary
- Contributing
- License
- Contacts
The PyRawS repository includes the following directories:
-
quickstart: it contains some subdirectories including jupyter notebooks:
API demonstration: it contains a notebook to demonstrate PyRawS API.DB_creation: it contains a notebook to automatically create a database for a target dataset.geographical_distribution: it contains a notebook to diplay the geographical distribution of the events of a dataset into a map.
-
PyRawS: it contains the PyRawS package. The latter is made of the following subdirectories:
database: it contains various PyRawS database and other databases.raw: it includes theRaw_eventandRaw_granuleclasses used to model, respectively, Sentinel-2 Raw events and Sentinel-2 Raw granules.l1: it contains theL1_eventandL1_tilesclasses used to model, respectively, Sentinel-2 L1C events and Sentinel-2 L1C tiles.utils: it contains some utilities for the PYRAW package.
-
resources: it contains various resources (i.e., images for the README)
-
scripts_and_studies: it contains various scripts and code for different studies realized to create the
THRAWSdataset. In particular:coregistration_study: it contains utils used to perform the coregistration study and desing the coarse coregistration technique.dataset_preparation: it contains scripts and other files used to design theTHRAWSfiles (i.e., download data, select events, and others).hta_detection_algorithms: it contains custom and simplified implementation of various high-thermal-anomalies-dection algorithms, including 1 used to design theTHRAWSdataset.runscripts: it contains some runscripts and utils to crop Sentinel-2 L1C tiles and generate useful images and export tif.granules_filtering: it contains a script to run and [1] on cropped Sentinel-2 L1-C tiles, extract bounding boxes and map them to the correspondent Raw granules.download_thraws: it contains the utility for the download of the dataset THRAWS.
Before all, clone this repository. We suggest using git from CLI, execute:
git clone https://github.com/ESA-PhiLab/PyRawS
# For Linux, the installation is straightforward.
# You just need to run the following command from the main directory:
\bin\bash\ source pyraws_install.sh
# NB: Your data should be placed in the data directory in the main.
# To install the environment, we suggest to use anaconda.
# You can create a dedicated conda environment by using the `environment.yml` file by running the following command from the main directory:
conda env create -f environment.yml
# To activate your environment, please execute:
conda activate PyRawS
Create a file sys_cfg.py in the directory pyraws\pyraws, and add two variables as follows:
PYRAWS_HOME_PATH="Absolute path to the main pyraws directory.
DATA_PATH="Absolute path to the data directory. "
By default the data directory is located in PyRawS main directory.
To use PyRawS with docker, use one of the following methods.
docker pull sirbastiano94/pyraws:latest
Follow these steps:
- Clone the repository and build the docker image by running the following command from the main directory:
docker build -t pyraws:latest --build-arg CACHEBUST=$(date +%s) -f dockerfile .
- Run the docker image by executing:
docker run -it --rm -p 8888:8888 pyraws:latest
If you want to perform the coregistration study, you need to:
- clone the repository SuperGlue Inference and Evaluation Demo Script;
- rename the subdirectory
modelstosuperglue_models; - move
superglue_modelsintocoregistration_study.
Coregistration study results can be generated by using the pyraws_coregistration_study.ipynb notebook. The database coregistration_study_db.csv containing info on the discarded eruption events is used to generate results in the notebook.
Coregistration notebook results can be generated by using the pyraws_coregistration_profiling.ipynb notebook.
All the data used by PyRawS need to be located in a directory called data, containing all the different datasets. You can place the data directory where you want and update the DATA_PATH variable in the sys_cfg.py file with its path (please, refer to: Set-up for coregistration study).
By default the data directory is located in PyRawS main directory.
To create a specific dataset, you need to create subfolder with the dataset name (e.g., THRAWS) in the data directory specific for your dataset. Such directory shall contain the following subdirectories:
raw: it will contain decompressed raw.l1c: it will contain L1C.
Every raw data and L1C data shall be contained in a specific subdirectory --placed in rawor l1c-- whose name shall match one of the ID_event entries of the .csv file, located in database directory.
To create the "THRAWS" database, the user should refer to the notebook called "database_creation.ipynb". This notebook contains the necessary code and instructions for creating the database. Simply follow the steps outlined in the notebook to successfully create the "THRAWS" database.
Please note that it is important to carefully follow the instructions in the notebook to ensure that the database is created correctly and without errors. Additionally, make sure that all required dependencies and packages are installed before running the code in the notebook.
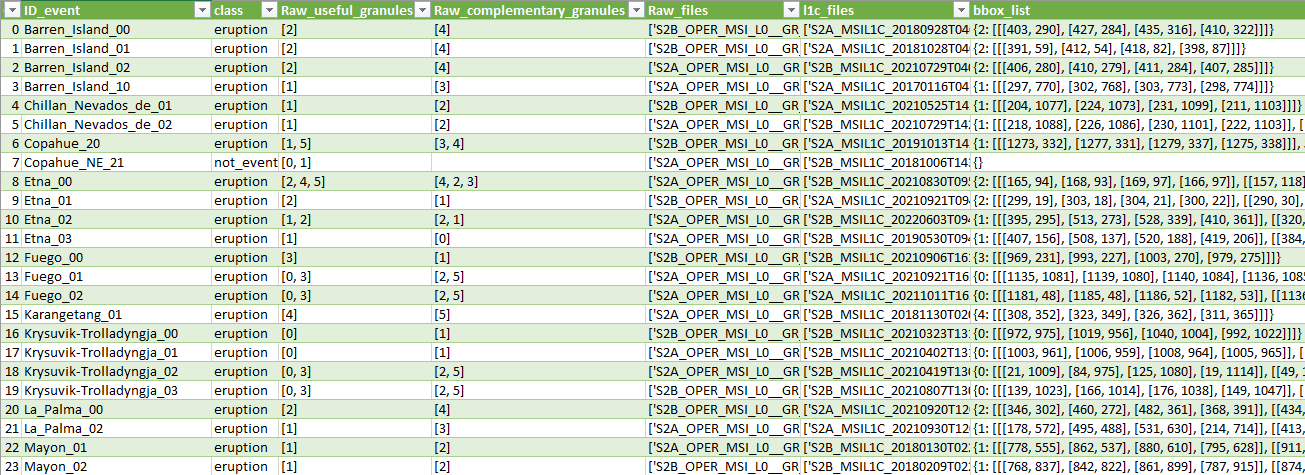
This respository contains an example of database (THRAWS) that is used by PyRawS to read and process Sentinel-2 Raw data and Sentinel-2 L1 data correctly. The minimal fields of the database include:
- ID_event: ID of the event (e.g., volcanic-eruption, wildfire, not-event). All the other fields of the row are referred to that
Sentinel-2acquisition. - class: class of the event (e.g., eruption, fire, not-event). Leave it empty
- Raw_useful_granules: list of Raw useful granules. Set to
Noneor leave it empty if you do not know what are the Raw useful granules. - Raw_complementary_granules: list of Raw complementry granules. Set to
Noneor leave it empty if you do not know what are the Raw complementry granules. - Raw_files: list of Raw granules (mandatory).
- l1c_files: list of L1 tiles (mandatory if you need L1C data).
- bbox_list: dictionary {Raw useful granules : [bounding box list for that granule]}. Set to
Noneor leave it empty if you do not know the bounding box location.
To create a new database (e.g., my_database_name), please, proceed as follows:
- Create a ".csv" file with the structure shown above and place it into the
databasesubfloders (e.g.,my_db.csv). You can use start from the thraws_db.csv database and edit it accordingly to your specification. - Create subdirectory
my_database_namein thedatasubdirectory and populate it with the correspondent Sentinel-2 Raw data and Sentinel-2 L1 data as described in Data directory. - Update the
DATABASE_FILE_DICTIONARYinPyRawS/utils/constants.pyas follows:
DATABASE_FILE_DICTIONARY={"THRAWS" : "thraws_db.csv", "my_database_name" : "my_db.csv"}
Now, you should be able to use your new database.
N.B The creation of a database is not mandatory. However, it is strongly advisable. Indeed, without creating a database you can still open Raw data as described in Open a Raw event from path. However, some pieces of information such as the Raw useful granules associated to a specific event, the event bounding boxes or the image class can be retrieved only when the database is set-up.
Downloading Sentinel-2 Raw data requires to specify a polygon surrounding the area of interest and a date. Given the pushbroom nature of the Sentinel-2 sensor, bands of data at Raw level do not look at the same area (i.e., they are not registered). Therefore,to be sure to collect all the band around an event (i.e., volcanic eruptions, wildfires) rectangular polygons centered on the events of area 28x10 B02) interesects the polygon area.
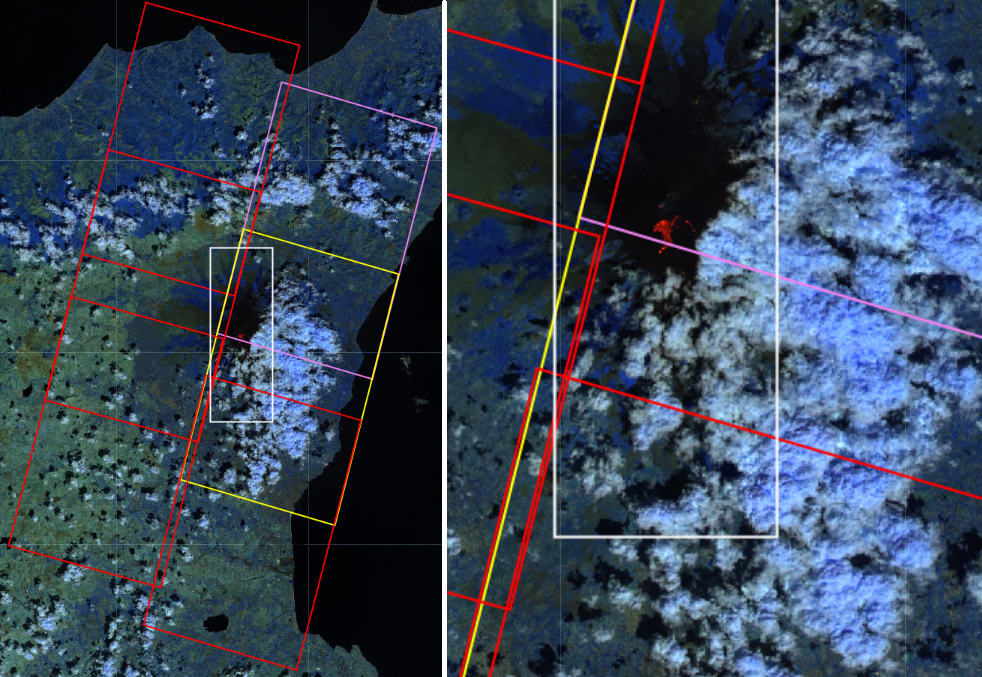
The image above shows the footprint of the all the Sentinel-2 Raw granules that are downloaded for the eruption named "Etna_00" in our database by using the white rectangular polygon. We define the collection of Raw granules that are downloaded for each of the rows of our database "Sentinel-2 Raw event".
However, as you can see in the image above, most of the Sentinel-2 Raw granules in Etna_00 Sentinel-2 Raw event do not contain the volcanic eruption (big red spot) of interest (red rectangulars). Indeed, only the yellow and the pink rectangulars intersects or include part of the volcanic eruption.
In addition, the fact that one Raw granule intersects or include one event, this does not mean that the latter interesects or is included in all the bands of that Raw granule. In particular,
since we use the bands [B8A, B11, B12] to detect wildfires and volcanic eruptions, we consider Raw useful granules those granules whose band B8A interesects the event. This is true for the yellow rectangular but not for the pink one (you need to trust us here, since the bands are not displaced in the image above). We take the band B8A only because after the coregistration, the other bands will be moved to match the area of B8A.
Finally, for some Raw useful granules part of the eruptions or the wildfire could extend until the top or the bottom edge of the polygon. In this case, some of the bands could be missing for a portion of the area of interest. To be sure that this is not happening, in addition to the Raw useful granules, it is important to consider Raw complementary granules, which fills the missing part of the interest bands of the Raw useful granules.
For each Sentinel-2 Raw event, the THRAWS dataset clearly states those Raw granules that are Raw useful granules or Raw complementary granules. However, the entire Raw granules collection is provided for each Raw event to permit users that wants to use other bands to detect warm temeprature anomalies to do it.
The next examples (with the exception of Open a Raw event from path) will exploit the THRAWS database to showcase the use of PyRawS, but they are applicable to any databases compatible with PyRawS.
The next code snipped will showcase how to use PyRawS to open a Raw_event My_Raw_data, included in the THRAWS database by using its PATH. We assume to have the My_RAW_data directory in the same directory where you execute the code snippet below.
To manipulate Raw events, PyRawS offer a class called Raw_event. To open an avent, we will use the Raw_event class method from_path(...), which parses the database specified, retrieves the event specified by id_event and opens it with the requested bands (bands_list).
When you open an event, you can specify which bands to use. If bands_list is not specified, the method from_path(...) will return all the bands.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B8A", "B11", "B12"]
#Read "Etna_00" from THRAWS
raw_event.from_path(#Path to the Etna_00 event
raw_dir_path="Path_to_my_RAW_data",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True)The example above can be used directly, even if you did not set up a PyRawS database. However, PyRawS offer some API to parse directly Raw_event parsing a database. with no need to specify a path. Please, check Open a Raw event from database.
The next code snipped will showcase how to use PyRawS to open the Raw_event Etna_00 included in the THRAWS database.
To do that,
To manipulate Raw events objects, PyRawS will exploits the Raw_event class method from_database(...), which parses the associated .csv file located in PyRawS/database with no need to specify the PATH from the user. To execute the next code snipped, we assume to you have already downloaded and set-up the THRAWS database as specificied in databases compatible with PyRawS.
As for the method from_path(...) described in Open a Raw event from path, you can specify which bands to use. If bands_list is not specified, the method from_database(...) will return all the bands.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B8A", "B11", "B12"]
#Read "Etna_00" from THRAWS
raw_event.from_database(#Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")All the next examples will assume you already have downloaded and set-up the THRAWS database as specificied in databases compatible with PyRawS. However, they can work by using from_path(...) instead of from_database(...) and specifying the PATH to the Etna_00 event manually.
As specified in Raw events and Raw granules, an Raw event is a collection of Raw granules. As for Raw_event, Raw granules are modelled in PyRawS through a dedicated class Raw_granule.
The next code snippet will show how to get the information about the Raw granules that compose the Etna_00 Raw_event. The class method show_granules_info() will print the list of events and some metada for each event. To get the same information as a dictionary {granule name : granule info} for an easy manipulation, you can use the Raw_event class method get_granules_info(...).
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B8A", "B11", "B12"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
# Printing granules info
raw_event.show_granules_info()
#Getting Raw granules info dictionary {granule name : granule info}
granules_info_dict=raw_event.get_granules_info()The class Raw_event contains a list of objects Raw_granule, each one modelling a specific Raw granule that belongs to that Raw_event (please, check Raw events and Raw granules for more information).
The different Raw_granule objects are sorted alphabetically and are accessible through indices. The next code snippet will show how to get a specific Raw granule by using the Raw_event class method get_granule(granule_idx), where granule_idx is the granule indices. The function returns an Raw_granule object. As for Raw_event objects, it is possible to print or retrieve metadata information for a spefic Raw_granule by using the Raw_granule methods get_granule_info() and show_granule_info().
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B8A", "B11", "B12"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
#Read the granule 0 of the Etna_00 event.
raw_granule_0 = raw_event.get_granule(0)
# Printing the info of the granule 0
raw_granule_0.show_granule_info()
#Getting Raw granules info dictionary {granule name : granule info}
granule_0_info_dict=raw_granule_0.get_granule_info()To visualize the values of an Raw_data object, it is possible to return it as a PyTorch tensor. However, since the different bands have different resolutions, depending on the bands that we want to shape as a tensor, it is necessary to upsample/downsample some of them to adapt them to the band with higher/smaller resolution. The next code snippet will open the Etna_00 with bands B02 (10 m), B8A (20 m), B11 (20 m), get the granule with index 1, and will return the first two bands in the collection as tensor by performing upsample.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
#Read the granule 1 of the Etna_00 event.
raw_granule_1 = raw_event.get_granule(1)
#Returning the bands B04 and B8A of raw_granule_1 as tensor by upsampling.
raw_granule_1_tensor=raw_granule_1.as_tensor(#list of bands to transform as tensor
requested_bands=["B04", "B8A"],
#Set to True to perform downsampling (default)
downsampling=False)It is possible to superimpose Raw_granule bands by using the class method show_bands_superimposition(...). (N.B. it is possible to superimpose up to three bands).
In case bands have different resolution, you need to specify if you want to superimpose them by performing downsampling (default) or upsampling. The next code snippet will open the Etna_00 with bands B02, B8A, B11, B12, get the granule with index 0, and will superimpose the last three bands.
from pyraws.raw.raw_event import Raw_event
import matplotlib.pyplot as plt
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11", "B12"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
#Read the granule 0 of the Etna_00 event.
raw_granule_0 = raw_event.get_granule(0)
#Returning the bands B04 and B8A of raw_granule_1 as tensor by upsampling.
raw_granule_0.show_bands_superimposition(#Bands to superimpose
requested_bands=["B04", "B11", "B12"],
#Set to True to perform downsampling
downsampling=True)
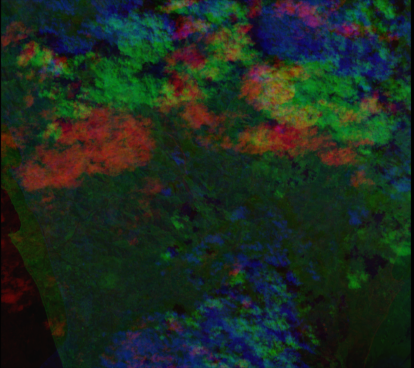
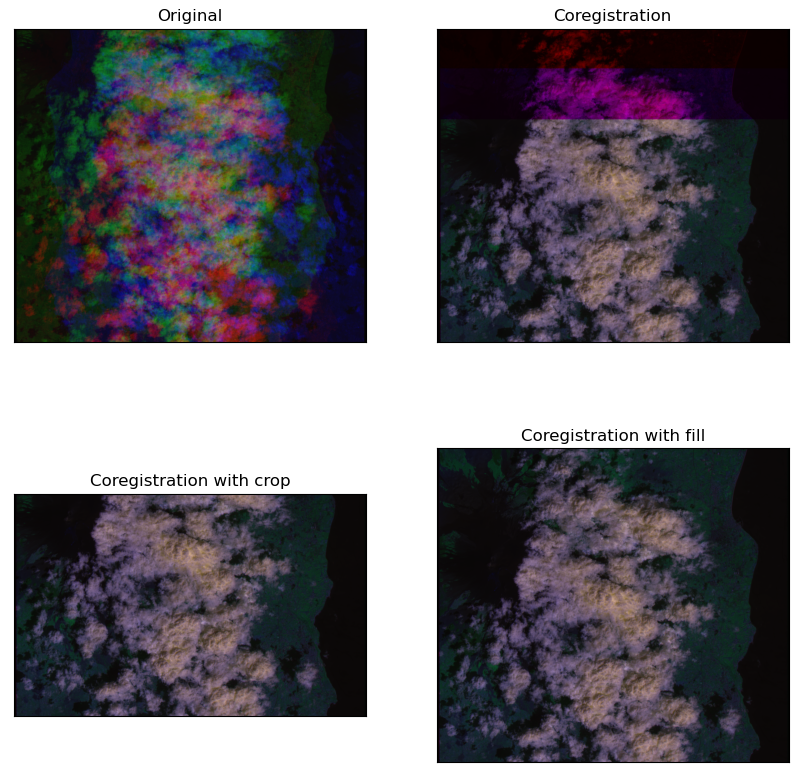
plt.show()The previous code snippet will display the image below. As you can see, the various bands of the image lack of coregistration of the various bands.
PyRawS offers some utils to perform coarse coregistration on Raw_granule objects. You can coregister a specific Raw_granule object of the Raw_event collection by calling the coarse_coregistration(...) method of the Raw_event class by selecting the correspondent index through the granules_idx input.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
# Perform the corase coregistration of the "Etna_00" event.
# Missing pixels will be filled with zeros.
raw_coreg_granule_2=raw_event.coarse_coregistration( # granule index to coregister.
granules_idx=[2])The previous code snippet returns the coarse coregistration of the granule 2 of the "Etna_00" event. The coarse coregistration is performed by shifting the bands B8A and B11 with respect to the band B04, which is the first in the collection. The missing pixels produced by the shift of the bands B8A and B11 will be filled by zeros. The superimposition of the coregistered bands with zero-filling is shown in the image below ("coregistration")
It is possible to launch crop the missing values by setting the argument crop_empty_pixels=True, as in the snippet below.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
# Perform the corase coregistration of the "Etna_00" event.
# Missing pixels will be cropped.
raw_coreg_granule_0_with_crop=raw_event.coarse_coregistration(# granule index to coregister.
granules_idx=[2],
# Cropping missing pixels.
crop_empty_pixels=True)Alternatively, you can fill the pixing pixels with filler elements taken from other Raw_granule objects when available. This is done by setting the argument use_complementary_granules=True. In this case, the compatibility of adjacent Raw_granule objects will be checked by the coarse_coregistration(...) API and use it in case it is available.
When filling Raw_granule objects are not available, missing pixels will be cropped if crop_empty_pixels is set to True. The superimposition of the coregistered bands with crop is shown in the image below ("coregistration with crop).
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
# Perform the corase coregistration of the "Etna_00" event.
# Missing pixels will be cropped.
raw_coreg_granule_0_with_fill=raw_event.coarse_coregistration(# granule index to coregister.
granules_idx=[2],
# Search for filling elements
# among adjacent Raw granules
use_complementary_granules=True,
# Cropping missing pixels
# when compatible Raw granules
# are not available
crop_empty_pixels=True)The superimposition of the coregistered bands with filling elements is shown in the image below ("coregistration with fill).
It is possible to get coordinates of the vertices of a Raw_granule object. Georeferencing is performed by using the information of the bands shift used to perform the coarse coregistration with respect to the band B02 and by exploiting the coordinates of the Raw granule footprint (please, refer to Sentinel-2 Products Specification Document). The code snippet below shows ho to get the information of the differnet bands of an Raw granule.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#You can also read it by using raw_event.from_path(...).
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
#Read the granule 1 of the Etna_00 event.
raw_granule_1 = raw_event.get_granule(1)
#Get bands coordinates
bands_coordinates_dict=raw_granule_1.get_bands_coordinates()The code snipped above returns a bands_coordinates_dict, a dictionary structured as {band_name : [BOTTOM-LEFT(lat, lon), BOTTOM-RIGHT(lat, lon), TOP-RIGHT(lat, lon), TOP-LEFT(lat, lon)]}.
This example will show you how to extract database metadata associated to an Raw event. To run the next example it is necessary to have set up database how described in databases compatible with PyRawS.
Database metadata include:
- Event class (e.g.,
eruption,fire,not_event) - List of Raw useful granules
- {Useful granule : Bounding box} dictionary
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#raw_event.from_path(...) cannot be used.
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
#Extract event class.
raw_event.get_event_class()
#Extract indices of Raw useful granules
raw_event.get_useful_granules_idx()
#Get {Useful granule : Bounding box} dictionary
raw_event.get_bounding_box_dict()This example will shows how to export an Raw_granule to TIF files. To this aim, you need to provide the path to a target directory, which will contain a TIF file for each band.
from pyraws.raw.raw_event import Raw_event
#Instantiate an empty Raw_event
raw_event=Raw_event()
#Bands to open.
bands_list=["B04", "B8A", "B11"]
#Read "Etna_00" from THRAWS database.
#raw_event.from_path(...) cannot be used.
raw_event.from_database( #Database ID_EVENT
id_event="Etna_00",
#Bands to open. Leave to None to use all the bands.
bands_list=bands_list,
#If True, verbose mode is on.
verbose=True,
#Database name
database="THRAWS")
#Apply coarse coregistration to the Raw granule with index 0 and return it
raw_granule_0=raw_event.coarse_coregistration([0])
#Save folder path
output_tif_folder="raw_target_folder"
#Export a TIF file for each band.
raw_granule_0.export_to_tif(save_path=output_tif_folder)-
Lightweight spatial coregistration method optimized for onboard-satellite applications. It simply shifts the various bands of a fixed factor that depends only on the bands, the satellite and detector number.
-
Sentinel-2 data at
level-0(L0) are data that are transmitted to Ground from Sentinel-2 satellites. TheL0format is compressed to diminish downlink bandwidth requirements. For more information, refer to the Sentinel-2 Products Specification Document -
In the frame of this project, the Sentinel-2 Raw represents a particular product in the Sentinel-2 processing chain that matches a decompressed version of Sentinel-2 L0 data with additional metadata that are produced on ground. Once decompressed,
Sentinel-2 Raw dataare the data available on Ground that better emulate the one produced by Sentinel-2 detectors with the exception of the effects due to compression and onboard equalization, which are not compensated at this stage. Therefore,Sentinel-2 raw dataare those exploited in this project. For more information, refer to the Sentinel-2 Products Specification Document.
N.B: the nomenclatureraw dataand its location in the Sentinel-2 processing chain is specific for this project only. -
A
granuleis the image acquired by a Sentinel-2 detector during a single acquisition lasting 3.6 s. Granules are defined at L0 level. However, since the processing perfomed on the ground between L0 and raw data does not alter the image content (with the exception of the decompression process) but just provide additional metadata, granules are defined also at Sentinel-2 Raw level. Given the pushbroom nature of the Sentinel-2 sensor, bands do not look at the same area at Raw level. For more information, refer to the Sentinel-2 Products Specification Document -
Sentinel-2 Raw data are produced by decompressing Sentinel-2 L0 data. To download L0 data, it is necessary to specify one polygon that surrounds a particular area-of-interest. This leads to download all those Sentinel-2 Raw granules whose reference band intersects the specified polygon. Such collection is a
Raw-event. EachRaw-eventmatches one of theID_evententry of the database.
For eachRaw-event, we do not provide all the collection of Sentinel-2 Raw granules, but only the set of Raw data useful granules and Raw data complementary granules. For an intuitive example, please, check Raw events and granules. -
The
Level 1-C(L1C) is one format forSentinel-2data. To convert Sentinel-2 Raw data toL1Cdata, numerous processing steps are applied to correct defects, including bands coregistration, ortho-rectification, decompression, noise-suppression and other. For more information, refer to the Sentinel-2 Products Specification Document. -
Same concept for Sentinel-2 Raw events but applied on Sentinel-2 L1C data.
-
The
Sentinel-2 L1C tileis the minimumL1Cproduct that can be downloaded. -
Given a certain set of bands of interest
[Bx,By,...,Bz],Raw complementarey granulesare the granules adjacents at Raw-useful-granules that that can be used to fill missing pixels of[By,...,Bz]bands due to their coregistration with respecto the bandBx. For an intuitive example, please, check Raw events and granules. -
Given a certain set of bands of interest
[Bx,By,...,Bz], whereBxis the first band in the set, anRaw useful granuleis one of the collection of Sentinel-2 Raw granules that compose a Sentinel-2 Raw event whose bandBxinclude (or intersects) a certain area of interest (e.g., an eruption or an area covered by a fire). For an intuitive example, please, check Raw data events and granules.
The PyRawS project is open to contributions. To discuss new ideas and applications, please, reach us via email (please, refer to Contacts). To report a bug or request a new feature, please, open an issue to report a bug or to request a new feature.
If you want to contribute, please proceed as follow:
- Fork the Project
- Create your Feature Branch (
git checkout -b feature/NewFeature) - Commit your Changes (
git commit -m 'Create NewFeature') - Push to the Branch (
git push origin feature/NewFeature) - Open a Pull Request
Distributed under the Apache License.
Created by the European Space Agency
- Gabriele Meoni - G.Meoni@tudelft.nl (previously, wit ESA
$\Phi$ -lab) - Roberto Del Prete - roberto.delprete at ext.esa.int and unina.it
- Nicolas Longepe - nicolas.longepe at esa.int
- Federico Serva - federico.serva at ext.esa.int