Brain Tumor-Segmentation using U2Net
Background: The rate of people suffering from cancer is increasing day by day. Manual segmentation of brain tumors for cancer diagnosis from large volumes of MRI images generated in the clinical routine is a specialized, difficult and time-consuming task. Therefore, manual tumor segmentation is often fraught with intra-rater and inter-rater variability, resulting in imprecise boundary delimitation. Automated brain tumor image segmentation is needed.
Goal: Automatic brain tumor segmentation is an important task in medical image processing. Early diagnosis of brain tumors is important for clinical evaluation and treatment planning. U2-Net, a two-level nested U-structure architecture, was used for the brain tumor segmentation task in this repository.
Overview
Dataset
The dataset used in this project was provided from the public collections of the The Cancer Imaging Archive (TCIA). A dataset was created by combining 2 public collections.
- This dataset contains T2-weighted and post-contrast T1-weighted images for each of the 159 subjects. Segmentation of tumors in three axial slices that include the one with the largest tumor diameter and ones below and above are provided in NIfTI format.
For a detailed information about the dataset please refer to this site.
- This dataset contains skull-stripped and co-registered multimodal (i.e. T1, T1-Gd, T2, T2-FLAIR) magnetic resonance imaging (MRI) volumes and both computer-aided and manually-corrected segmentation labels in NIfTI format for each of the 65 subjects. T1-weighted and T2-weighted MRI were included in this study.
For a detailed information about the dataset please refer to this site.
Data Augmentation
The basic data augmentation methods listed below were used:
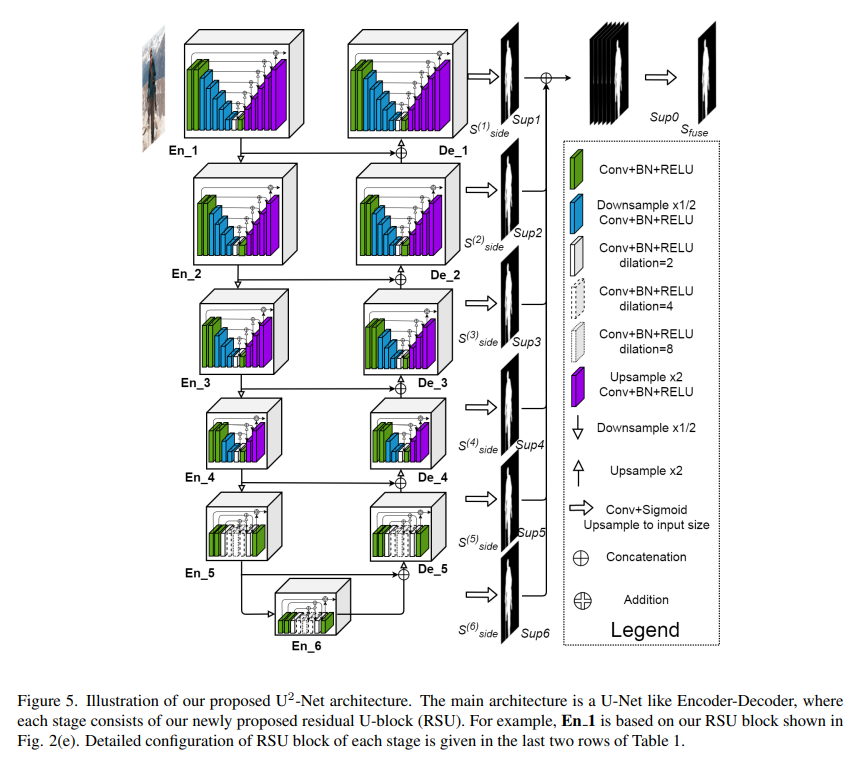
Model Architecture
Model architecture is shown in the figure below.
Training Process
The model was trained on Tesla P100 GPU offered by Google Colab Pro. Training took about 1 hour with U2NETP architecture and about 1 hour 30 minutes for U2NET architecture.
The graphs show epoch-dice loss (left) and epoch-loss (right) over 100 epochs are given below for train (blue) and validation (red).
The graphs also show evaluation dice loss (left) per iteration and loss per iteration (right) over 100 epochs are given below.
Metrics
To quantify the performance of our image segmentation, Dice Score is used. The algorithm is validated by calculating the Dice score, which is a measure of how similar the objects are. So it is the size of the overlap of the two segmentations divided by the total size of the two objects. That is, the score for a particular class c is the size of the overlap between the predicted region and its true counterpart.
Results
Some of the results are as follows.
| . | . |
|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |





