This project uses U-Net Architecture to create segmentation masks for brain tumor images.
Dataset used in this project was provided by Jun Cheng.
This dataset contains 3064 T1-weighted contrast-enhanced images with three kinds of brain tumor. For a detailed information about the dataset please refer to this site.
Version 5 of this dataset is used in this project. Each image is of dimension 512 x 512 x 1 , these are black and white images thus having a single channel.
Some Data Samples
| Original Image | Mask Image |
|---|---|
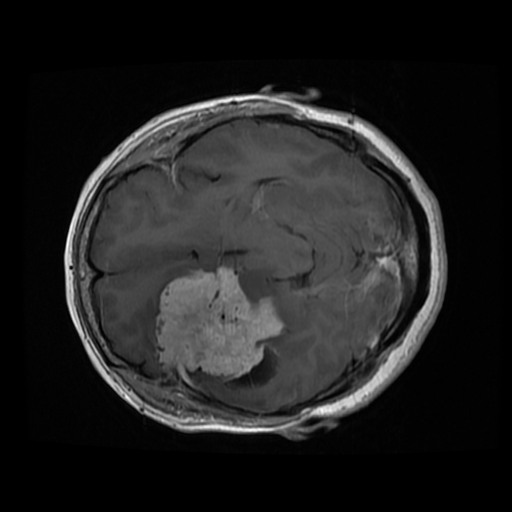 |
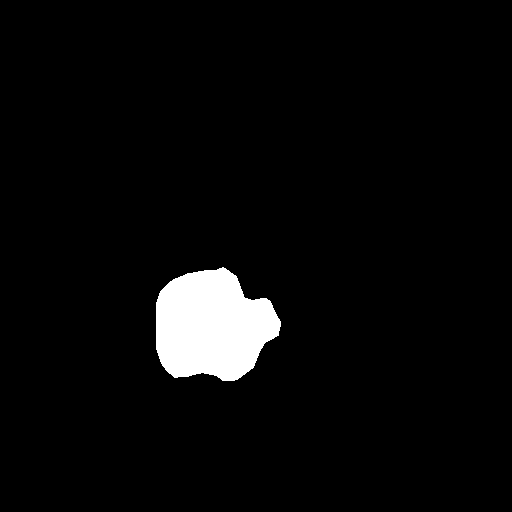 |
The basic forms of data augmentation are used here to diversify the training data. All the augmentation methods are used from Pytorch's Torchvision module.
- Horizontally Flip
- Vertically Flip
- Rotation Between 75°-15°
Code Responsible for augmentation
Each augmentation method has a probability of 0.5 and the order of application is also random. For Rotation Augmentation the degree of rotation is chosen randomly between 75°-15°.
The model architecture is depicted in this picture.
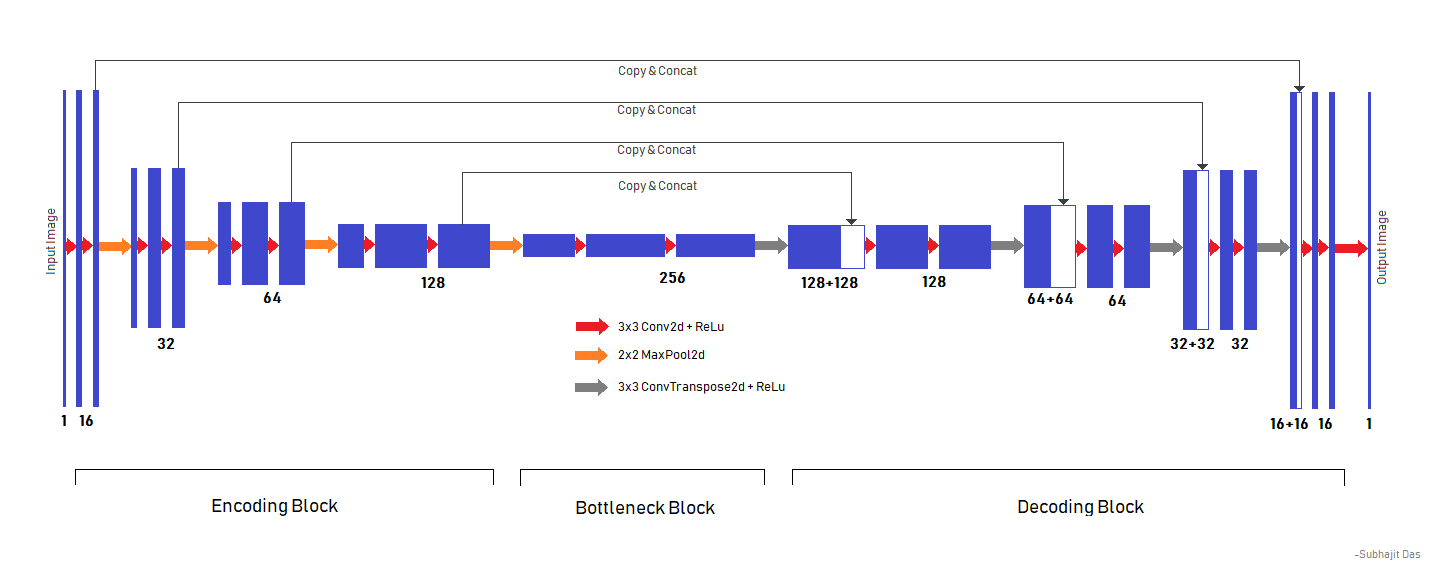
The model was trained on a Nvidia GTX 1050Ti 4GB GPU. Total time taken for model training was 6 hours and 45 minutes. We started with an initial learning rate of 1e-3 and reduced it by 85% on plateauing, final learning rate at the end of 100 epochs was 2.7249e-4.
Some graphs indicating Learning Rate & Loss Value over 100 epochs are given below.
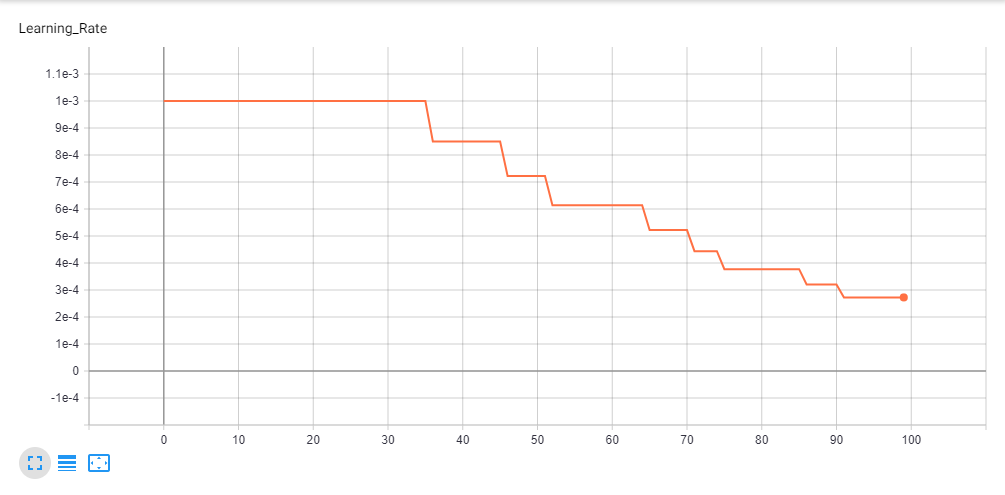 Learning Rate Graph in Tensorboard.
Learning Rate Graph in Tensorboard.
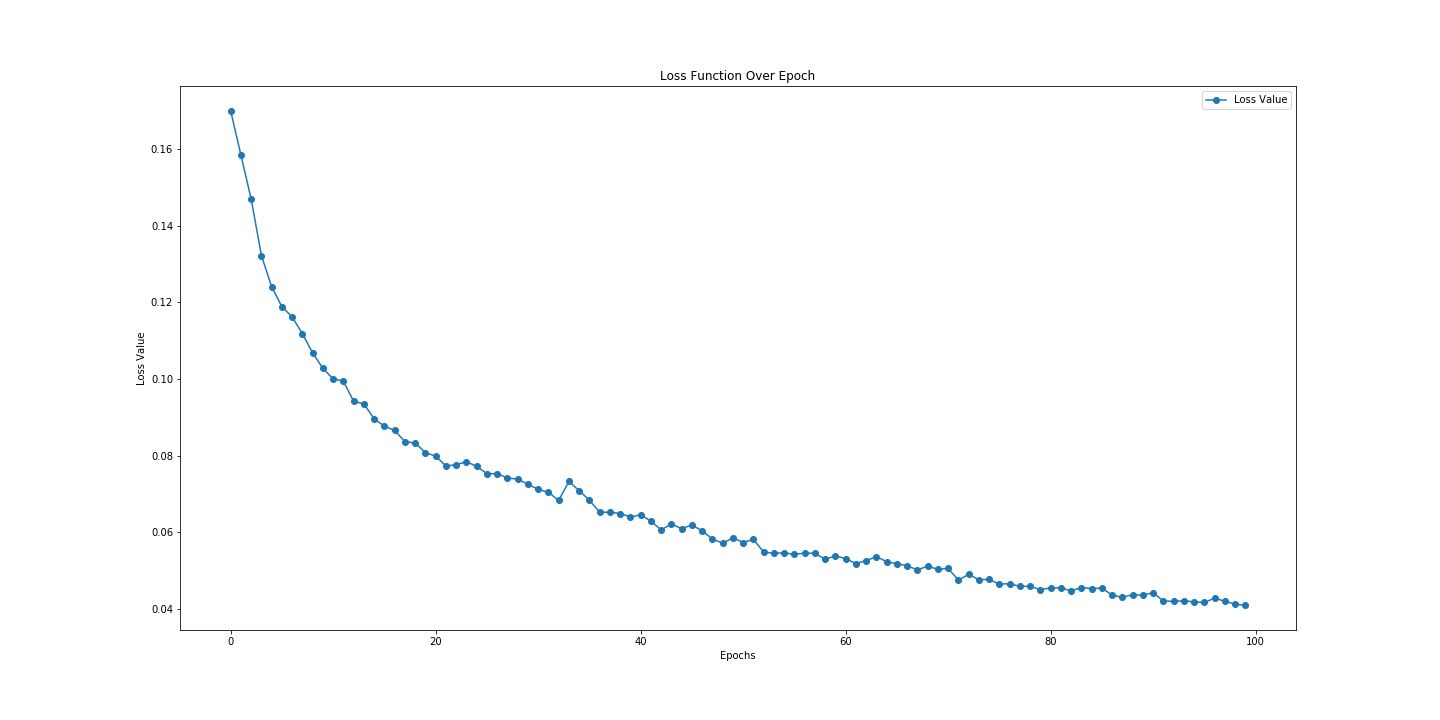 Loss Graph Plotted in Matplotlib
Loss Graph Plotted in Matplotlib
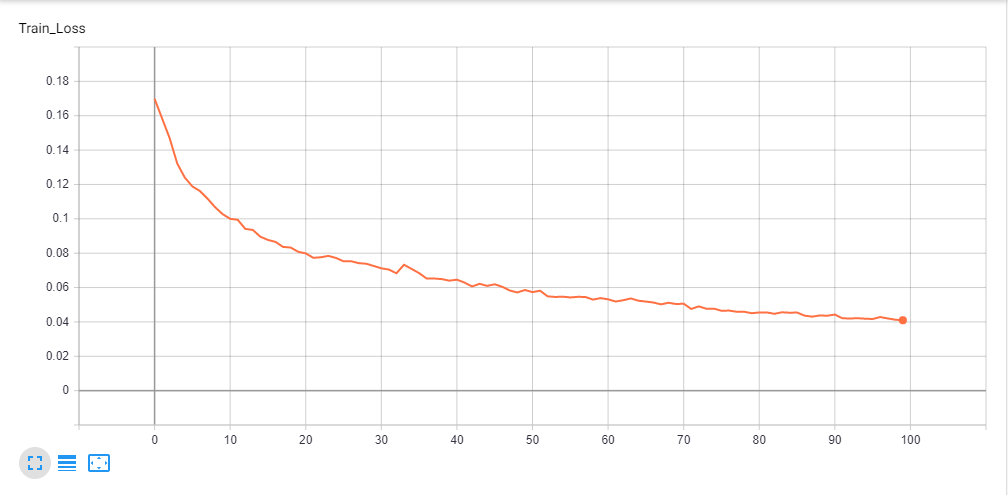 Loss Graph Plotted in Tensorboard
Loss Graph Plotted in Tensorboard
To see the complete output produced during the training process check this
This project uses python3.
Clone the project.
git clone https://github.com/Jeetu95/Brain-Tumor-Segmentation.gitInstall Pytorch from this link
Use pip to install all the dependencies
pip install -r requirements.txtTo open the notebook
jupyter labTo see logs in Tensorboard
tensorboard --logdir logs --samples_per_plugin images=100To setup the project dataset
python setup_scripts/download_dataset.py
python setup_scripts/unzip_dataset.py
python setup_scripts/extract_images.pypython api.py --file <file_name> --ofp <optional_output_file_path>
python api.py --folder <folder_name> --odp <optional_output_folder_path>
python api.py -hAvailable api.py Flags
--file : A single image file name.
--ofp : An optional folder location only where the output image will be stored. Used only with --file tag.
--folder: Path of a folder containing many image files.
--odp : An optional folder location only where the output images will be stored. Used only with --folder tag.
-h, --help : Shows the help contents.
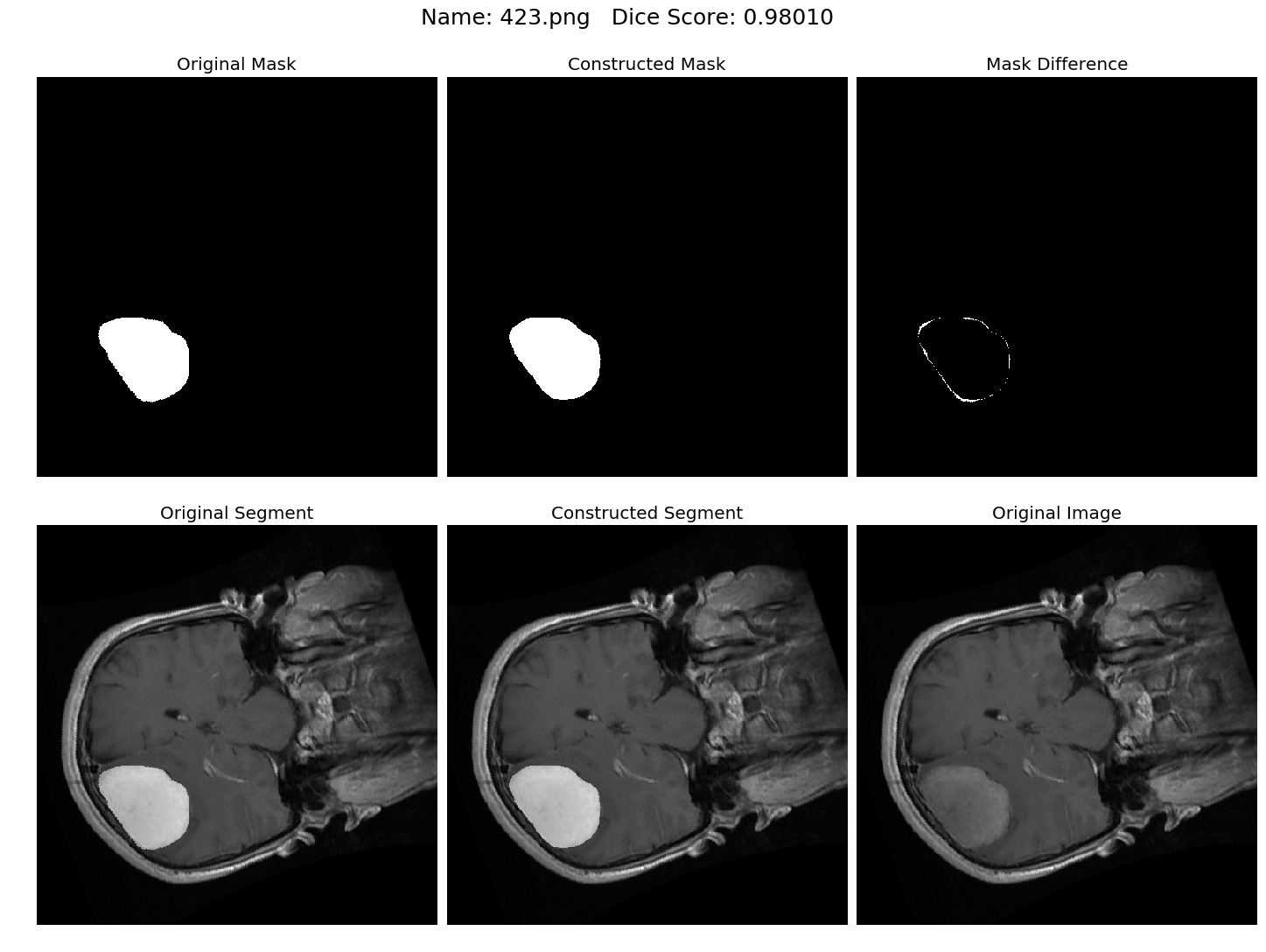
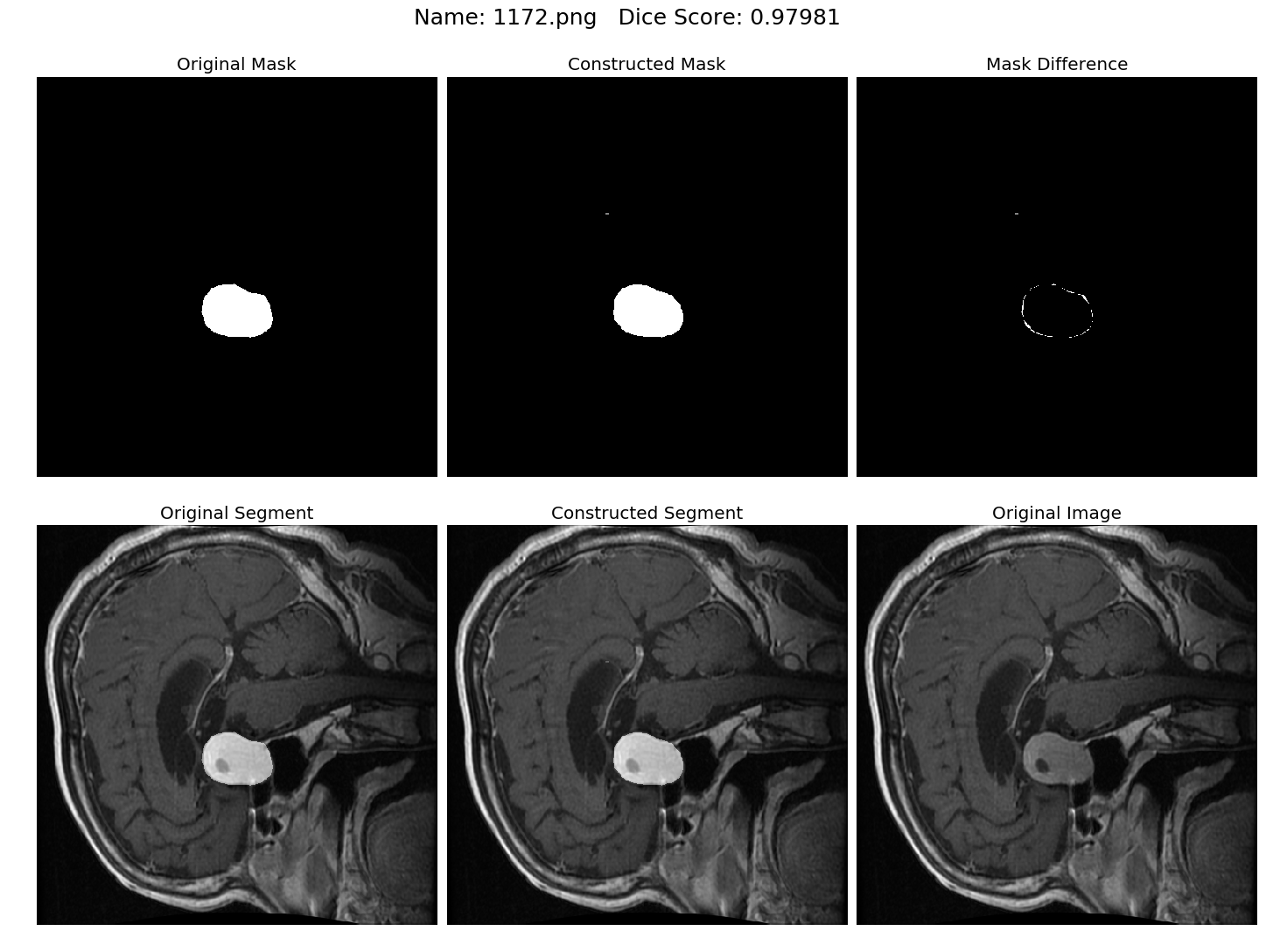
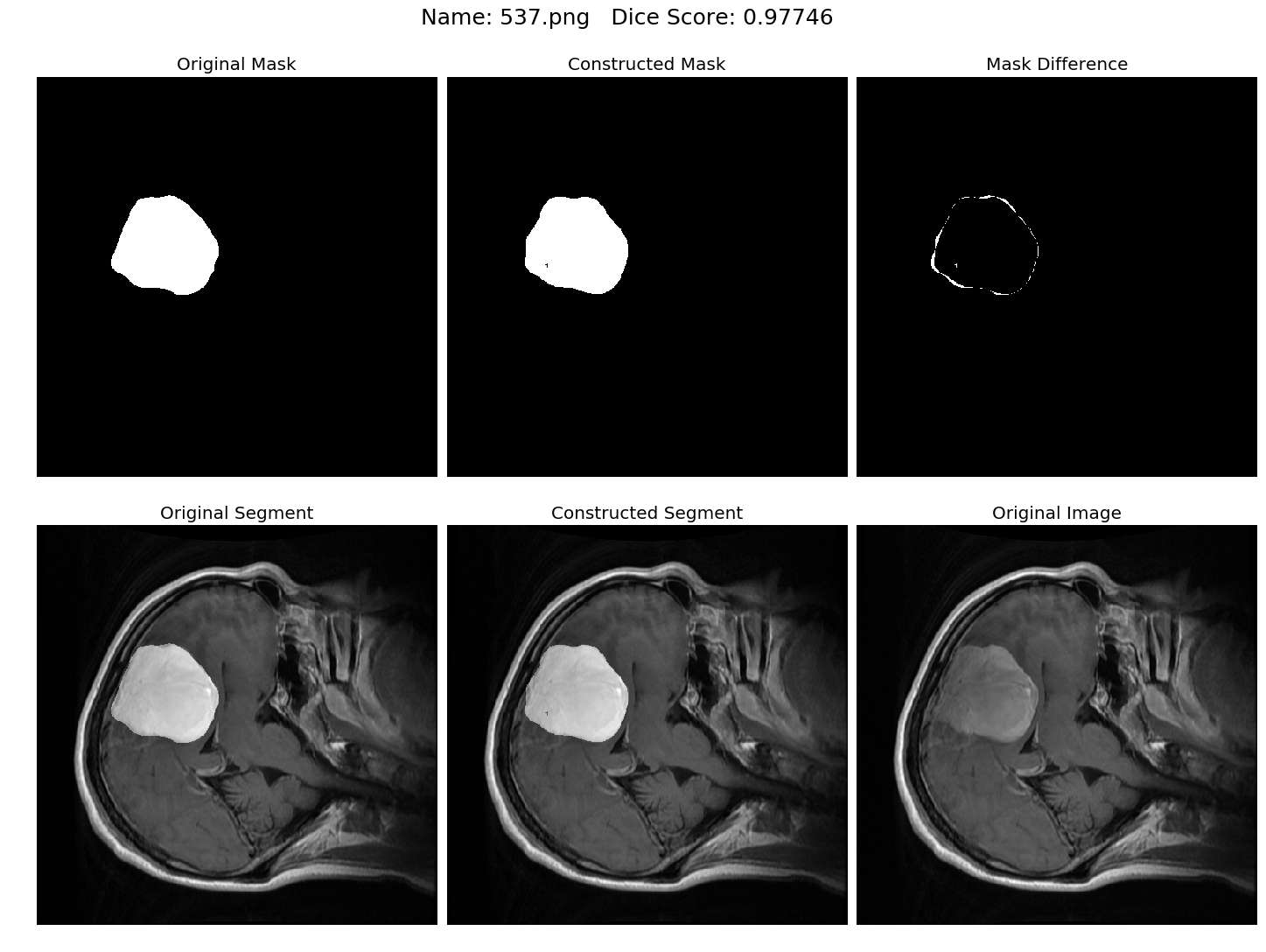
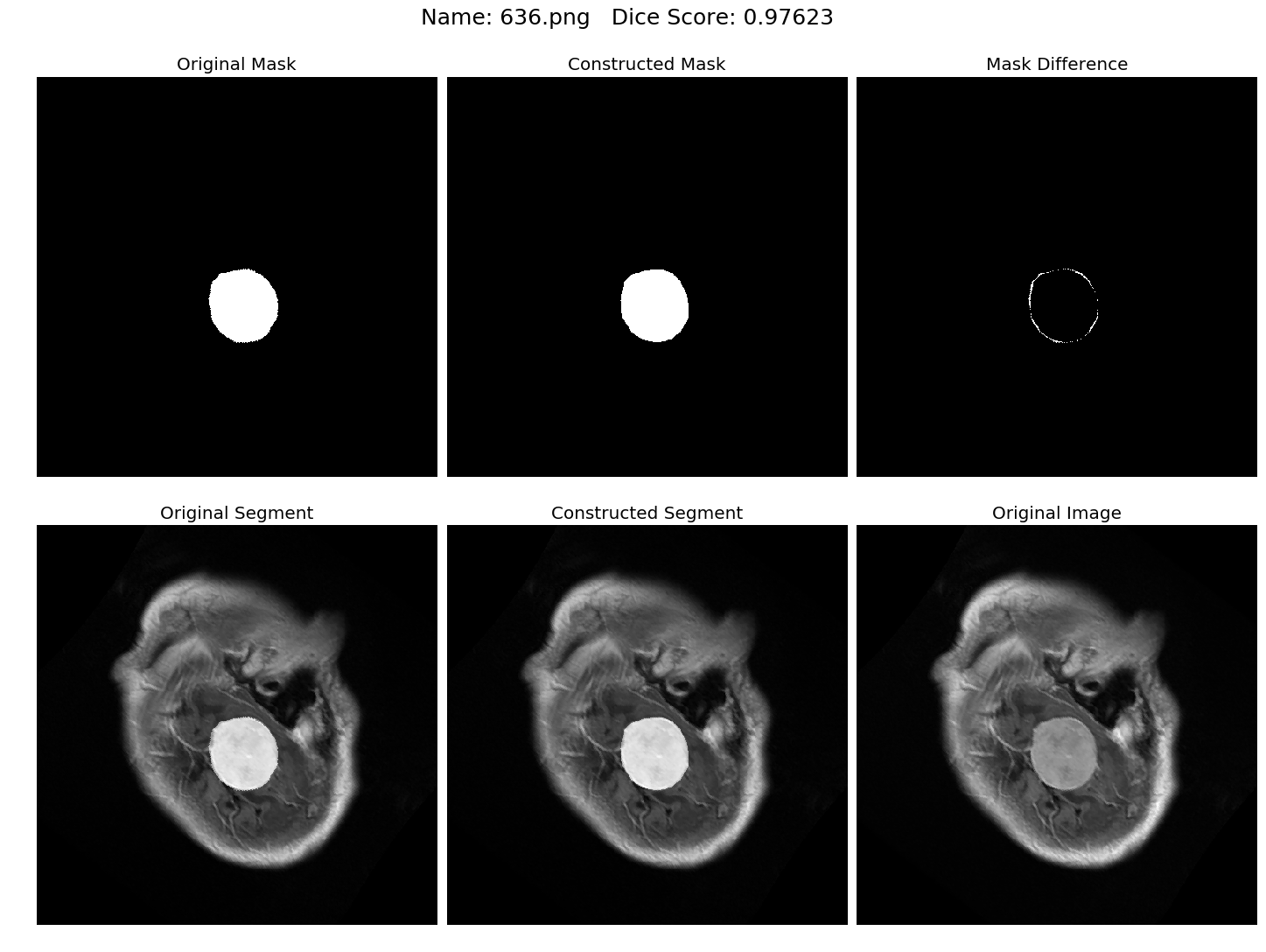
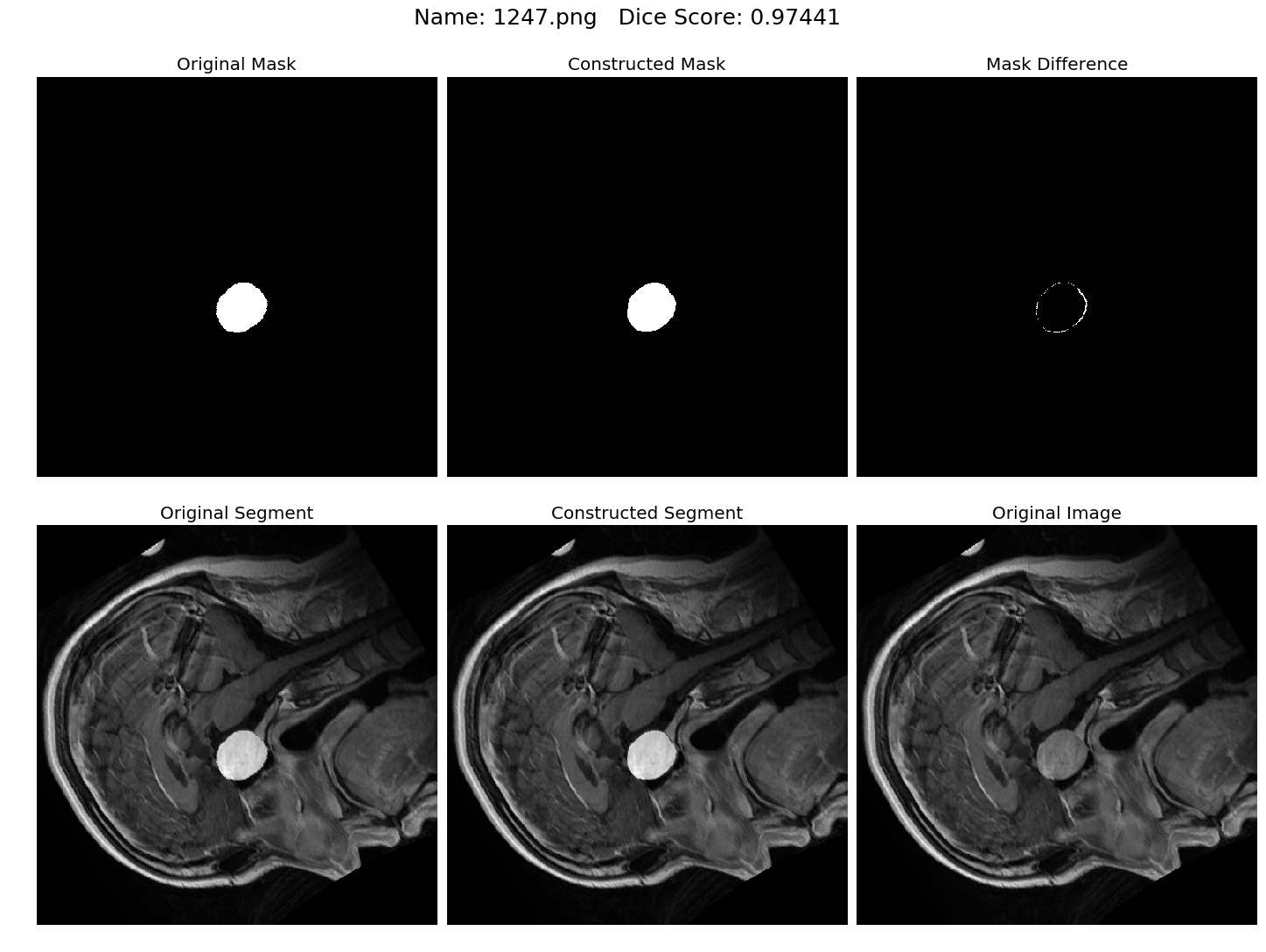
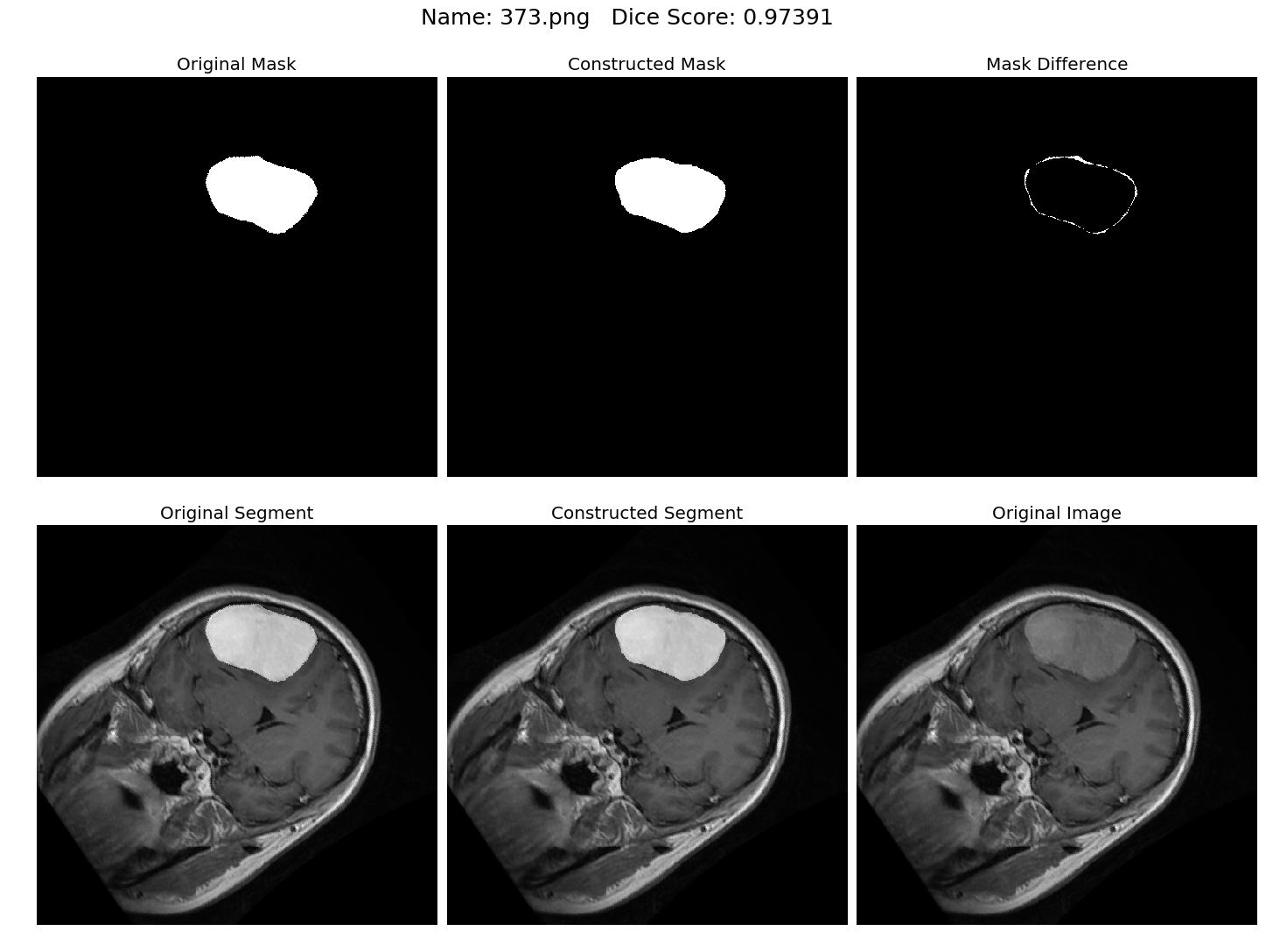
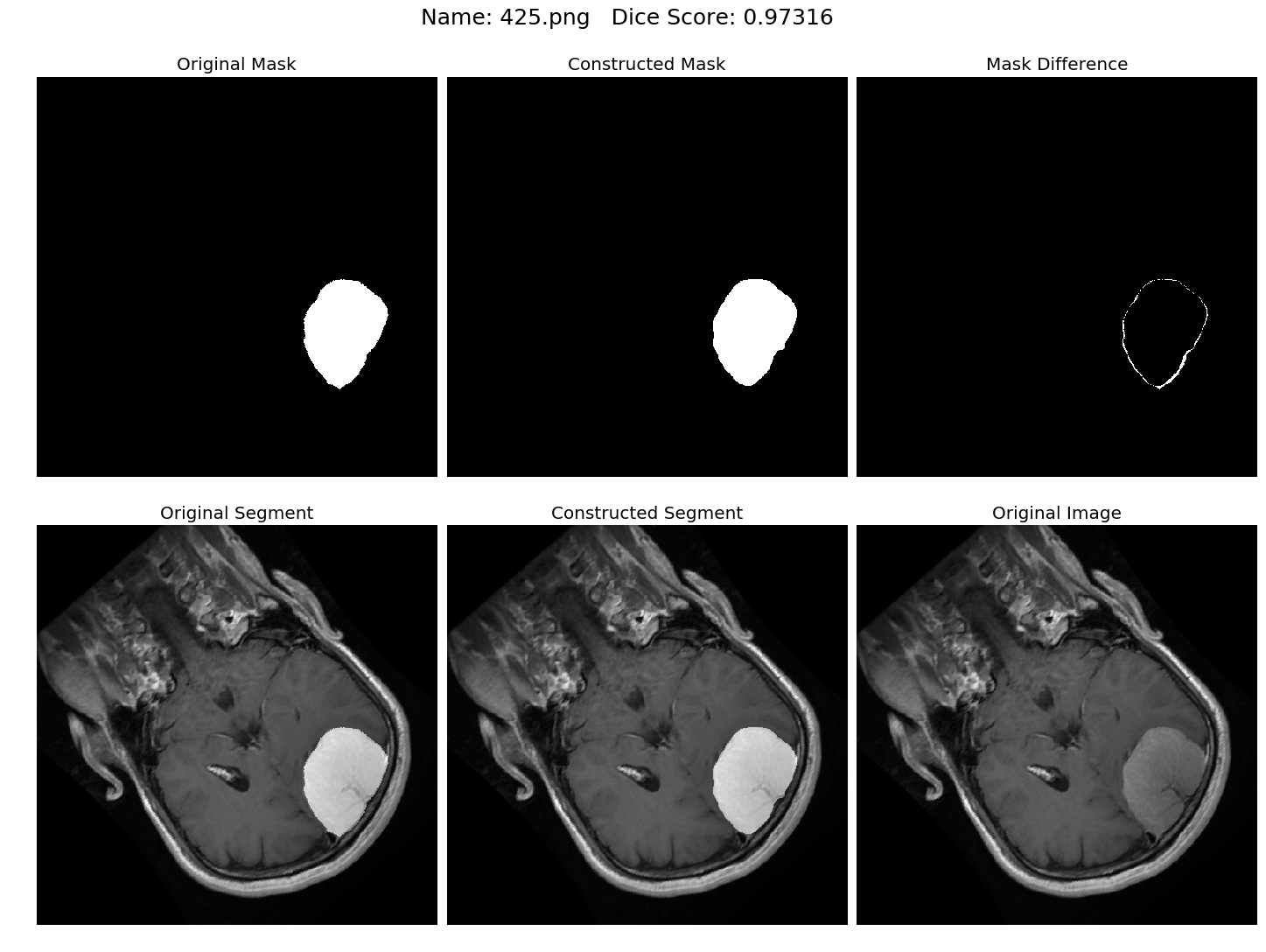
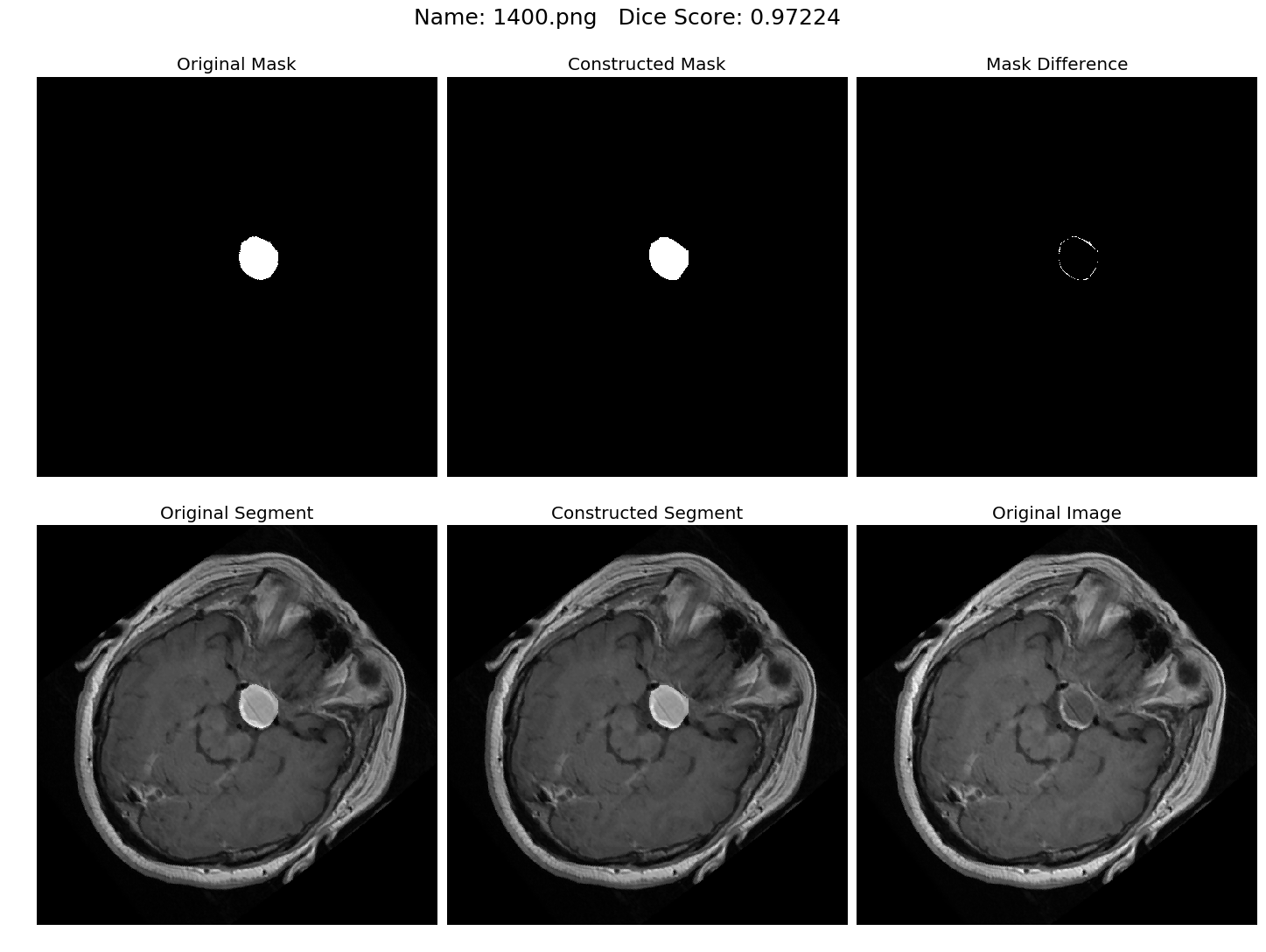
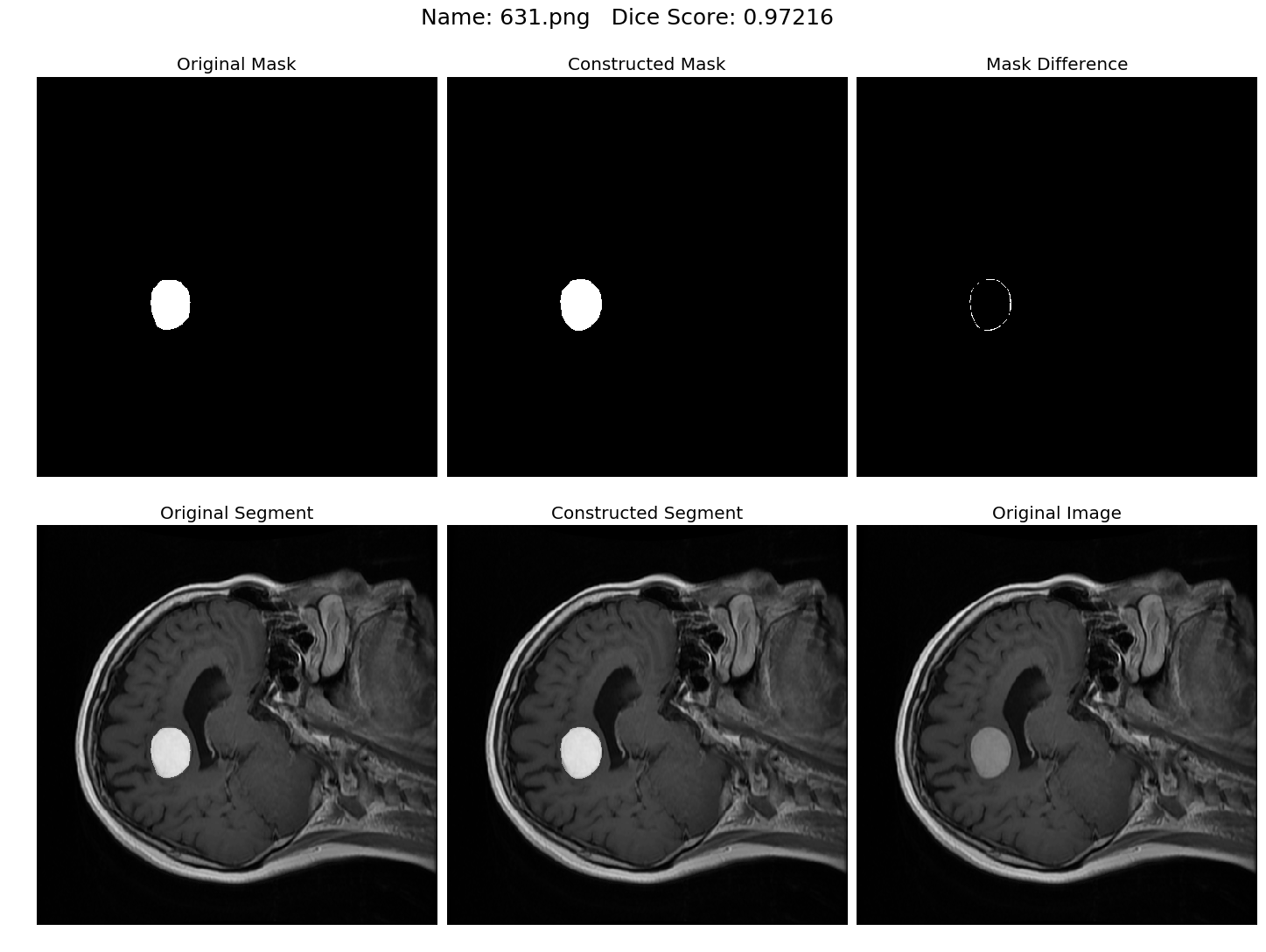
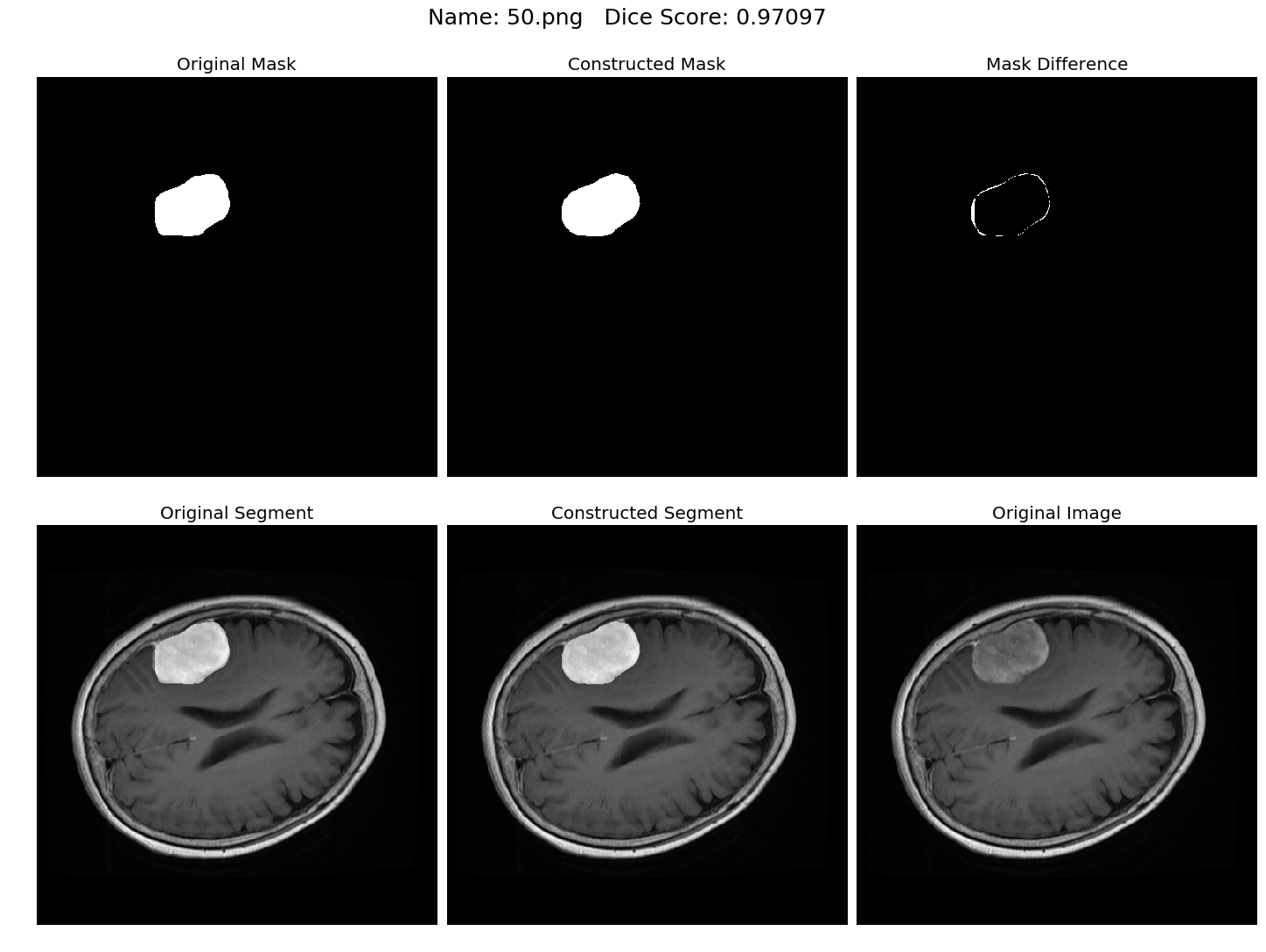
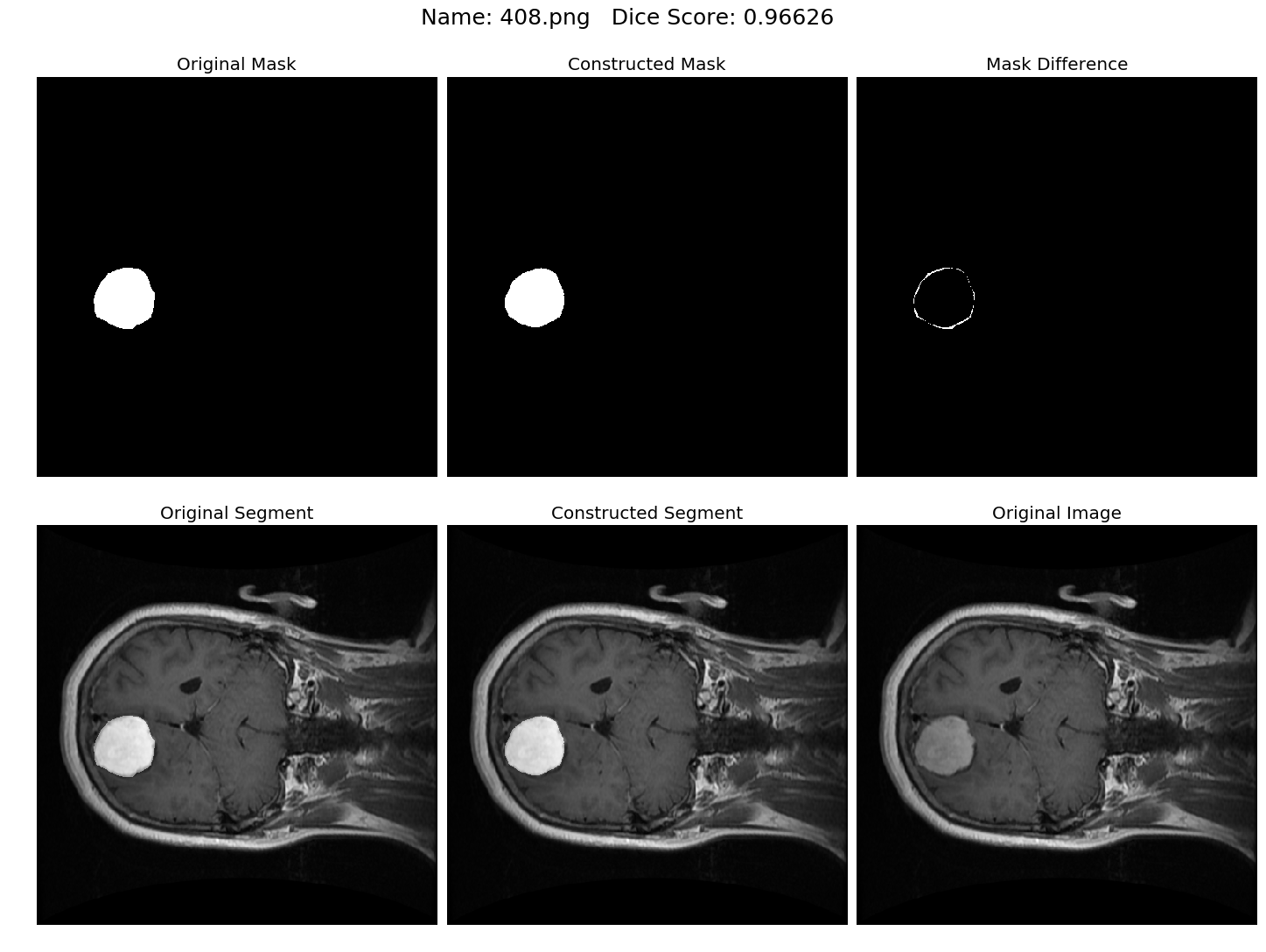
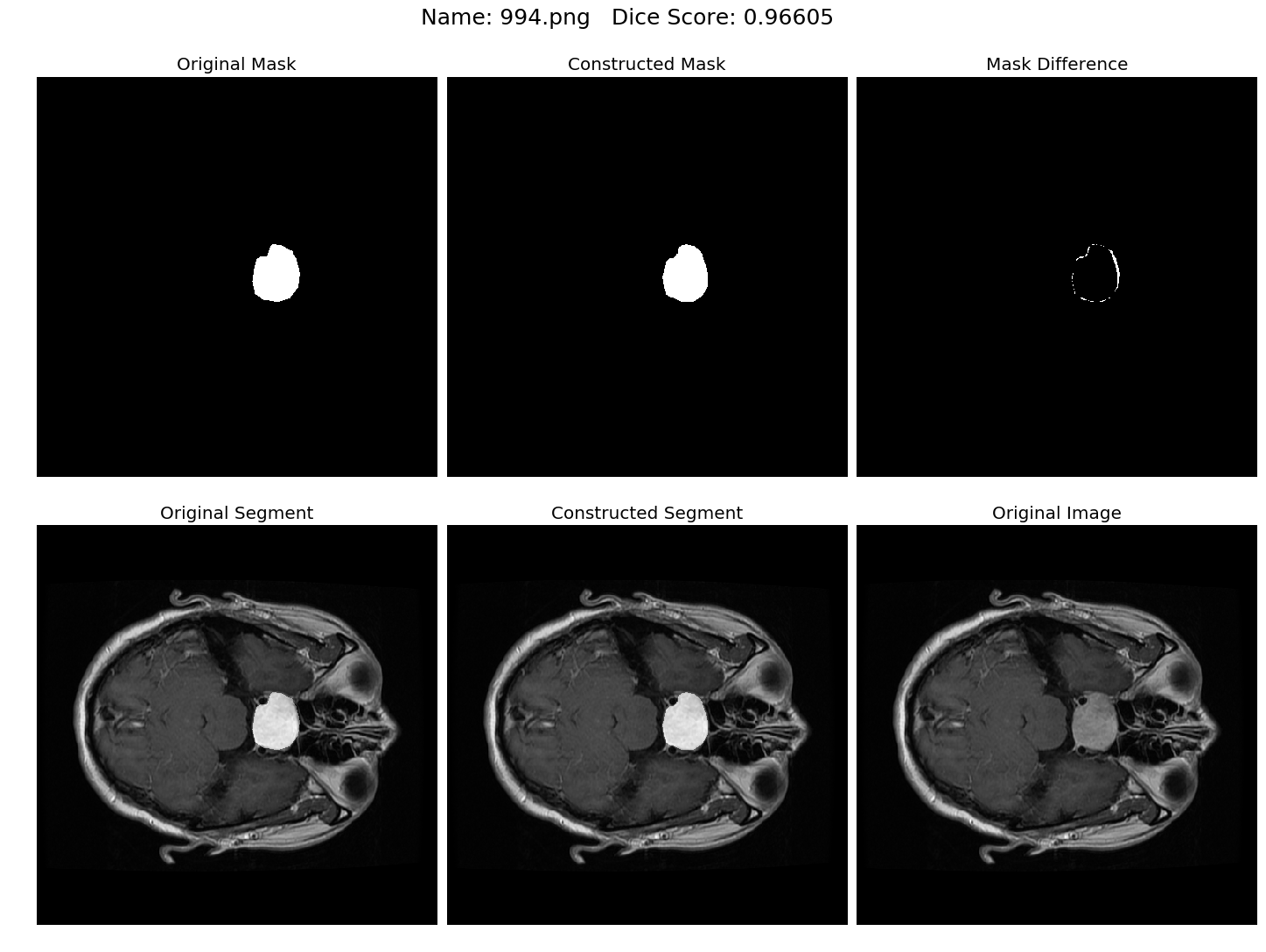
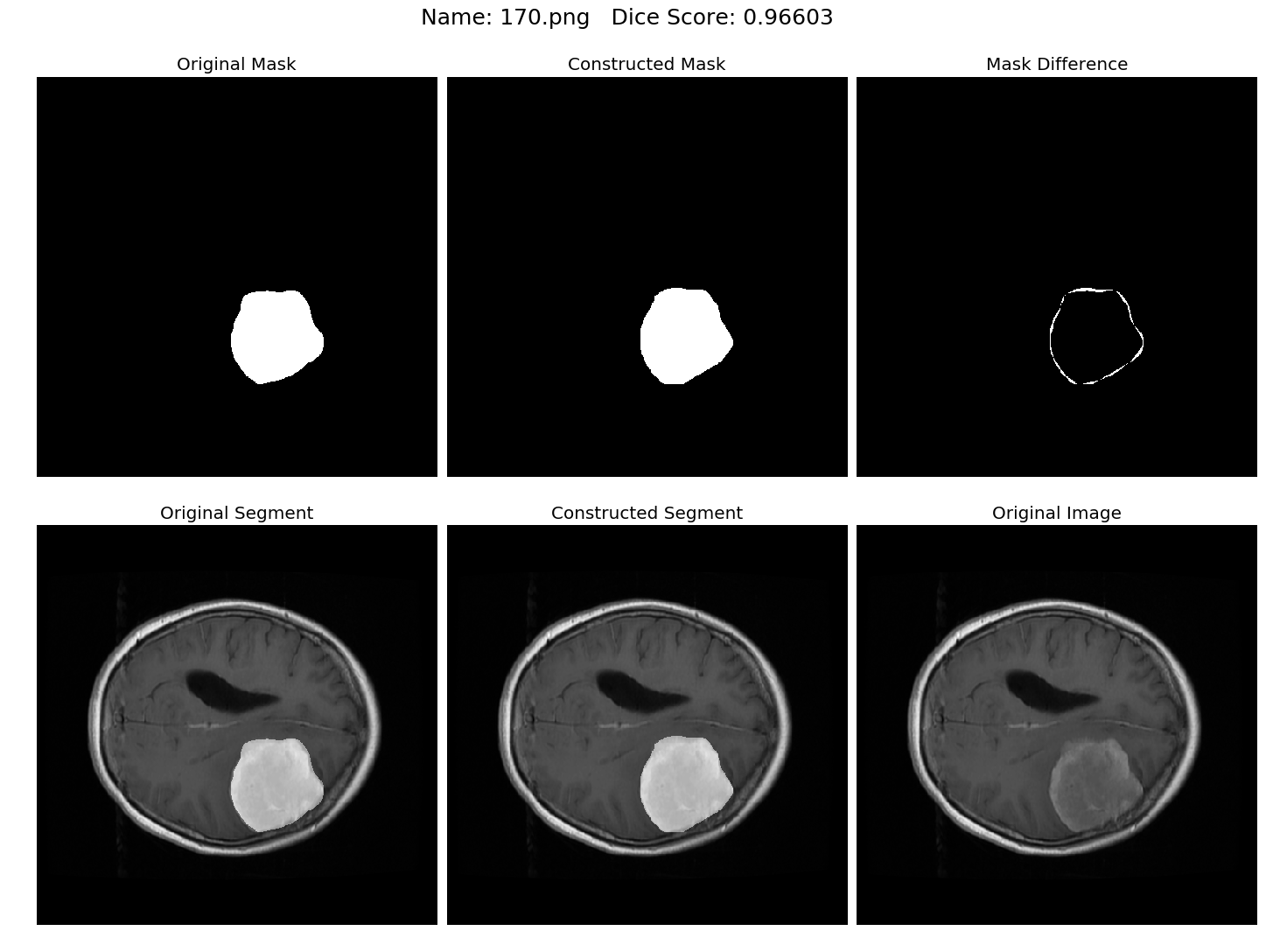
Some results generated by API are here
The mean Dice Score our model gained was 0.74461 in testing dataset of 600 images.
From this we can conclude that in our testing dataset our constructed mask has a similarity of about 74% with the original mask.
Some samples from our training dataset output are below. The top best results are here.To see all the results click on this Google Drive link
| . | . |
|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
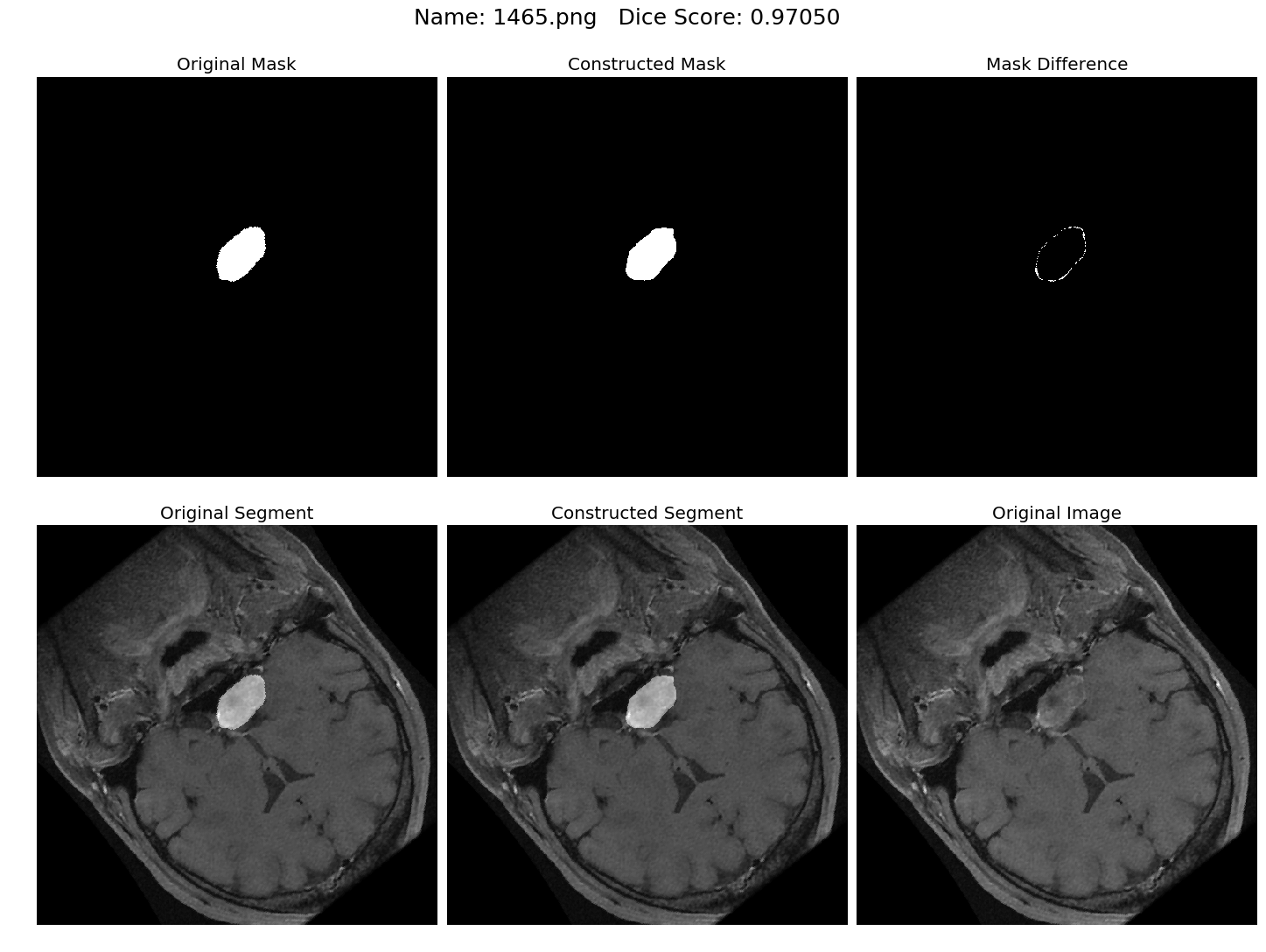 |
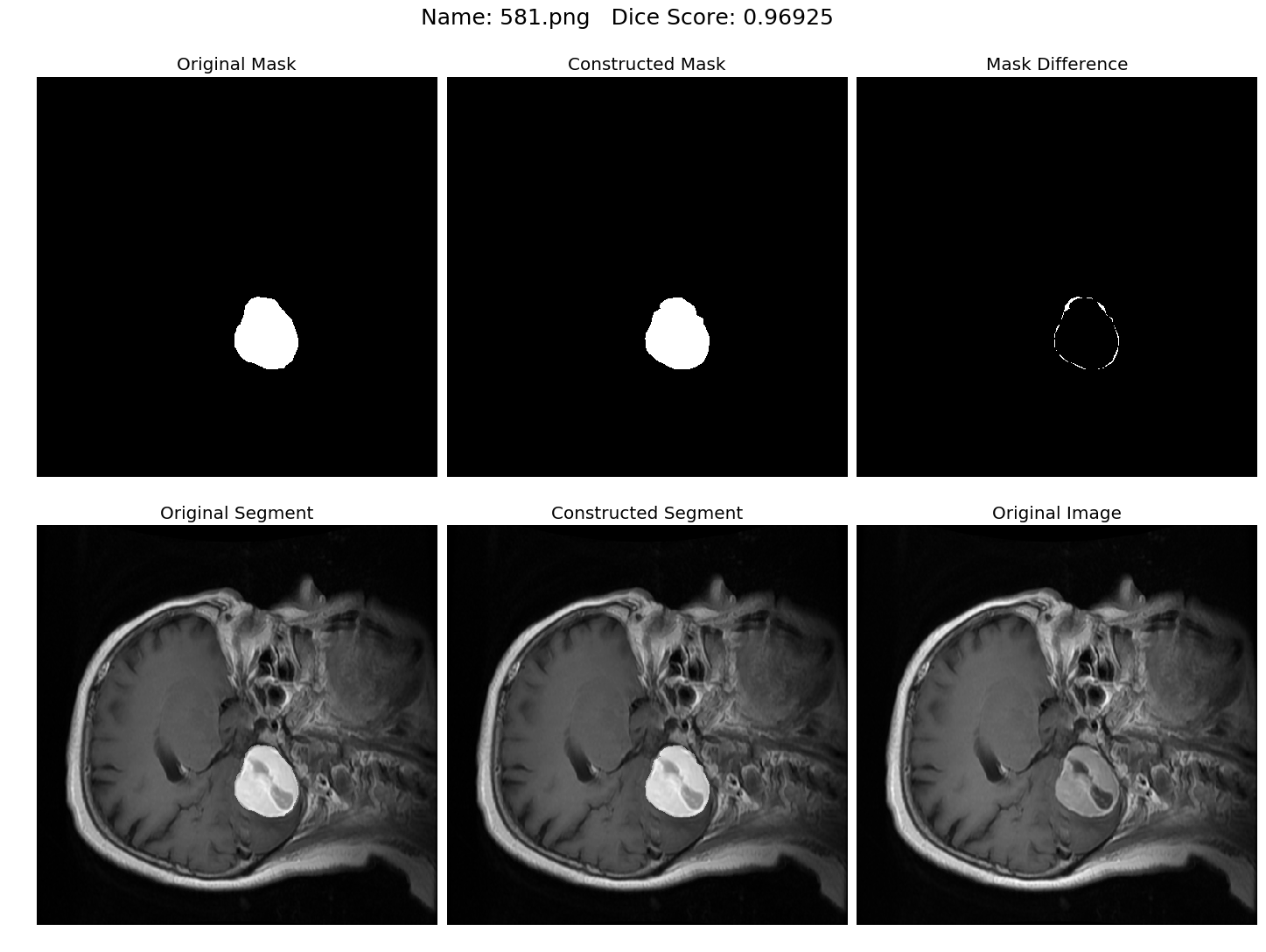 |
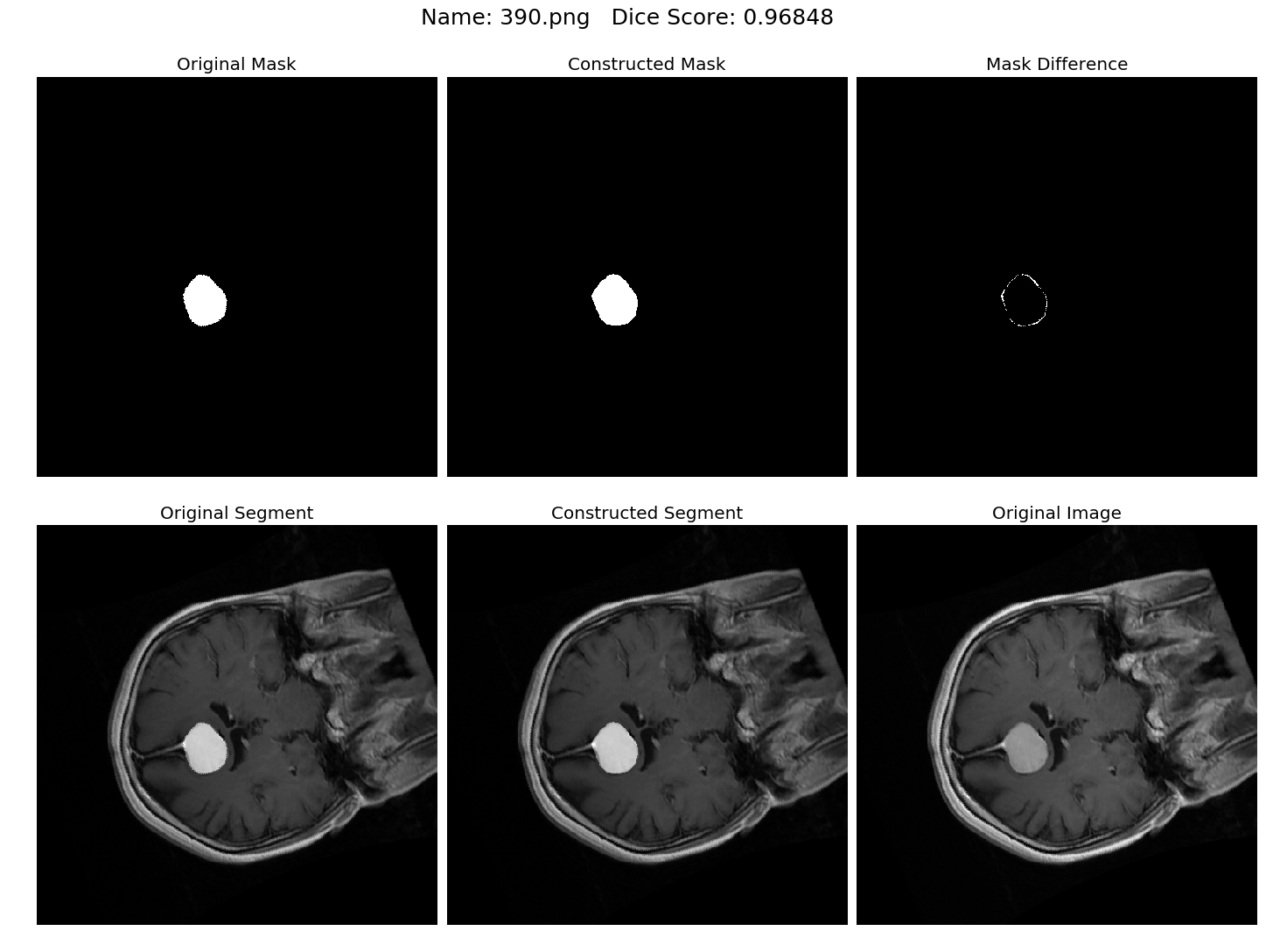 |
 |
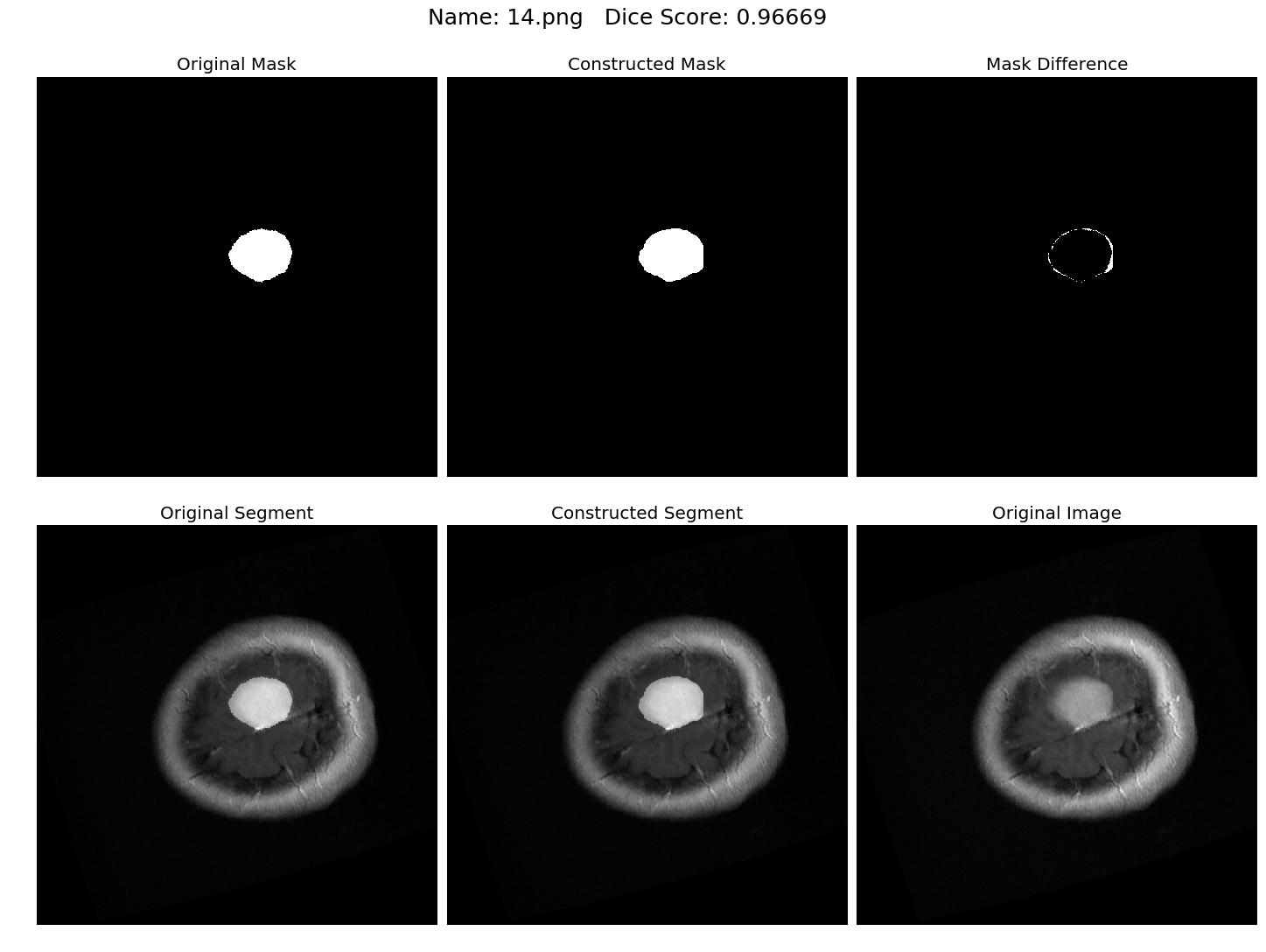 |
 |
 |
 |
 |
 |
Pull requests are welcome. For major changes, please open an issue first to discuss what you would like to change.







