MetaFX
MetaFX (METAgenomic Feature eXtraction) is an open-source library for feature extraction from whole-genome metagenome sequencing data and classification of groups of samples.
The idea behind MetaFX is to introduce the feature extraction algorithm specific for metagenomics short reads data. It is capable of processing hundreds of samples 1-10 Gb each. The distinct property of suggest approach is the construction of meaningful features, which can not only be used to train classification model, but also can be further annotated and biologically interpreted.
MetaFX documentation is available on the GitHub wiki page.
Here is a short version of it.
Old version of MetaFX is now deprecated and archived.
Table of contents
Idea of MetaFX
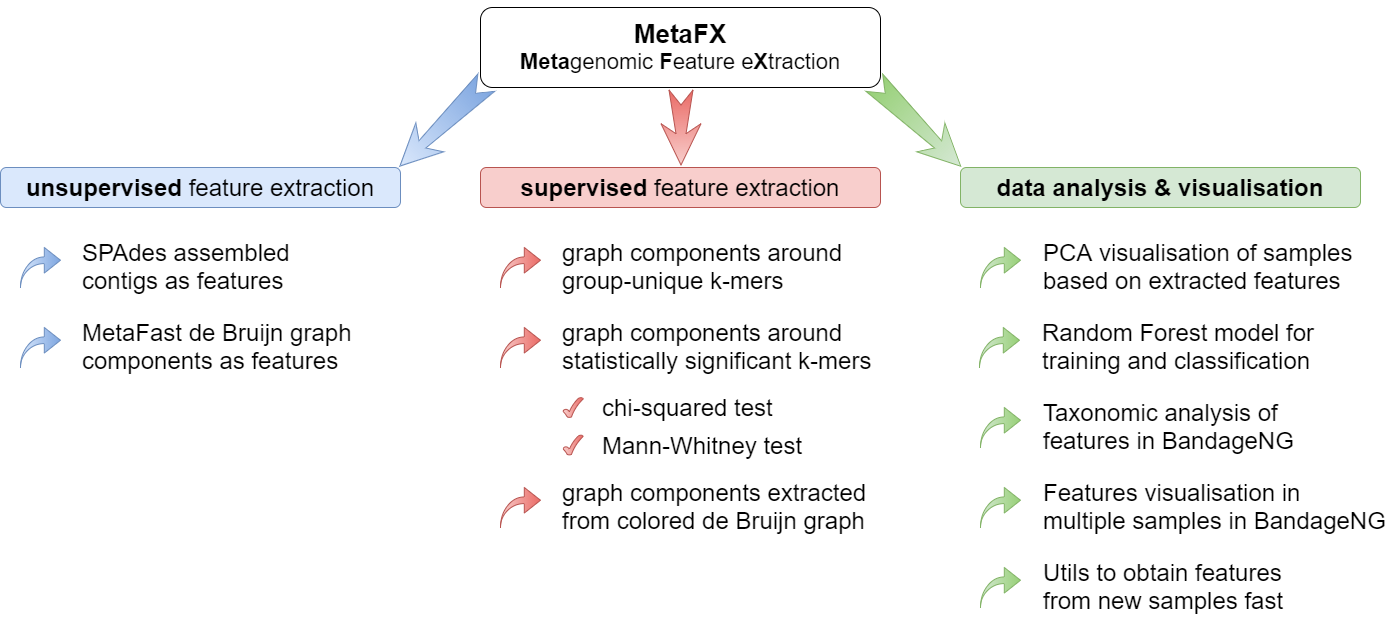
MetaFX is a toolbox with a lot of modules divided into three groups:
Unsupervised feature extraction pipelines
There are pipelines aimed to extract features from metagenomic dataset without any prior knowledge about samples and their relations. Algorithms perform (pseudo-)assembly of samples separately and construct the de Bruijn graph common for all samples. Further, graph components are extracted as features and feature table is constructed.
Supervised feature extraction pipelines
There are pipelines aimed to extract group-relevant features based on metadata about samples such as diagnosis, treatment, biochemical results, etc. Dataset is split into groups of samples based on provided metadata information and group-specific features are constructed based on de Bruijn graphs. The resulting features are combined into feature table.
Methods for classification and interpretation
There are pipelines for analysis of the feature extraction results. Methods for samples similarity visualisation and training machine learning models are implemented. Classification models can be trained to predict samples' properties based on extracted features and to efficiently process new samples from the same environment.
Installation
To run MetaFX, one need to clone repo with all binaries.
git clone https://github.com/ctlab/metafx
cd metafxThen add MetaFX binary directory to the PATH variable.
export PATH=/path/to/metafx/bin:$PATHFor permanent use, add the above line to your ~/.profile or ~/.bashrc file.
Requirements:
- JRE 1.8 or higher
- python=3.9.5
- python libraries listed in
requirements.txtfile. Can be installed using pip
python -m pip install --upgrade pip
pip install -r requirements.txt- coreutils required for macOS (e.g.
brew install coreutils) - If you want to use
metafx metaspadespipeline, you will also need SPAdes software. Please follow their installation instructions (not recommended for first-time use).
Scripts have been tested under Ubuntu 18.04 LTS, Ubuntu 20.04 LTS, macOS 11 Big Sur, and macOS 12 Monterey, and should generally work on Linux/macOS.
Multiple cores can be used to speed up computations.
Required RAM grows linearly with the size of the input dataset. Hard drive space for intermediate computations and results also growth linearly. For example, to process 12GB dataset in tutorial we used 16GB disk space, 8GB RAM, and 6 threads, which took 1 hour to process.
Running instructions
To run MetaFX use the following syntax:
metafx <pipeline> [<Launch options>] [<Input parameters>]To view the list of supported pipelines run metafx -h or metafx --help.
To view help for launch options and input parameters for selected pipeline run metafx <pipeline> -h or metafx <pipeline> --help.
MetaFX supports both single-end and paired-end input files. For correct detection of paired-end reads, files should be named with suffixes "_R1"&"_R2" or "_r1"&"_r2" after sample name before extension. For example, sample_r1.fastq&sample_r2.fastq, or reads_R1.fq.gz&reads_R2.fq.gz.
By running MetaFX a working directory is created (by default ./workDir/).
All intermediate files and final results are saved there.
Video tutorial
Details about installation and first use of MetaFX are available in the next video on youtube:
Examples
Examples and documentation for all MetaFX modules can be found in the Wiki.
Here is presented a minimal example of data analysis with MetaFX algorithms:
Step 1. Extract features from samples of three categories
metafx unique -t 2 -m 1G -w wd_unique -k 31 -i test_data/sample_list_train.txtInput parameters
| parameter | description |
|---|---|
| -t <int> | number of threads to use |
| -m <MEM> | memory to use (values with suffix: 1500M, 4G, etc.) |
| -w <dirname> | working directory |
| -k <int> | k-mer size (in nucleotides) |
| -i <filename> | tab-separated file with 2 values in each row: <path_to_file>\t<category> |
Output files
| file | description |
|---|---|
| wd_unique/categories_samples.tsv | tab-separated file with 3 columns: <category>\t<present_samples>\t<absent_samples> |
| wd_unique/samples_categories.tsv | tab-separated file with 2 columns: <sample_name>\t<category> |
| wd_unique/feature_table.tsv | tab-separated numeric features file: rows – features, columns – samples |
| wd_unique/contigs_<category>/seq-builder-many/sequences/component.seq.fasta | contigs in FASTA format as features for each category (suitable for annotation and biological interpretation) |
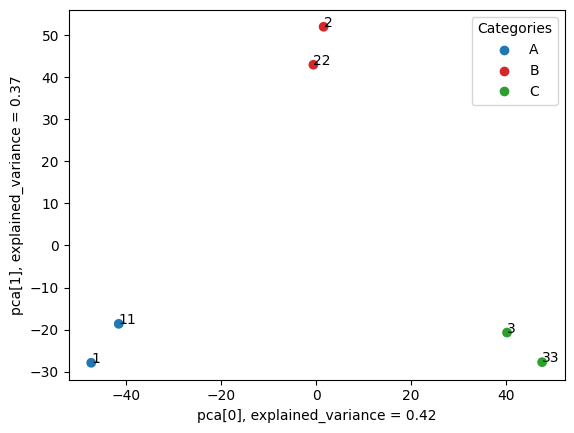
Step 2. Visualise samples proximity
metafx pca -w wd_pca -f wd_unique/feature_table.tsv -i wd_unique/samples_categories.tsv --showInput parameters
| parameter | description |
|---|---|
| -w <dirname> | working directory |
| -f <filename> | file with feature table in tsv format: rows – features, columns – samples |
| -i <filename> | tab-separated file with 2 values in each row: <sample>\t<category> |
| --show | print samples' names on plot |
Output files
wd_pca/pca[.png|.svg] – PCA visualisation of samples based on extracted features. As a result you should obtain the similar image showing the clear separation of samples into three clusters.
Step 3. Train classification model for category prediction
metafx cv -t 2 -w wd_cv -f wd_unique/feature_table.tsv -i wd_unique/samples_categories.tsv -n 2 --gridInput parameters
| parameter | description |
|---|---|
| -t <int> | number of threads to use |
| -w <dirname> | working directory |
| -f <filename> | file with feature table in tsv format: rows – features, columns – samples |
| -i <filename> | tab-separated file with 2 values in each row: <sample>\t<category> |
| -n <int> | number of folds in cross-validation |
| --grid | perform grid search of optimal parameters for classification model |
Output files
wd_cv/rf_model_cv.joblib – trained Random Forest model to predict samples' categories based on extracted features.
Step 4. Process new samples with hidden categories
metafx calc_features -t 2 -m 1G -w wd_new_samples -k 31 -d wd_unique/ \
-i test_data/test_A_R1.fastq.gz test_data/test_A_R2.fastq.gz \
test_data/test_B_R1.fastq.gz test_data/test_B_R2.fastq.gz \
test_data/test_C_R1.fastq.gz test_data/test_C_R2.fastq.gzInput parameters
| parameter | description |
|---|---|
| -t <int> | number of threads to use |
| -m <MEM> | memory to use (values with suffix: 1500M, 4G, etc.) |
| -w <dirname> | working directory |
| -k <int> | k-mer size (in nucleotides) |
| -d <dirname> | directory with results from MetaFX feature extraction module, containing folders with components.bin file for each category |
| -i <filenames> | list of reads files from single environment (FASTQ, FASTA, gzip- or bzip2-compressed) |
Output files
wd_new_samples/feature_table.tsv – tab-separated numeric features file for new samples: rows – features, columns – samples.
Step 5. Get prediction results for new samples
metafx predict -w wd_predict -f wd_new_samples/feature_table.tsv --model wd_cv/rf_model_cv.joblibInput parameters
| parameter | description |
|---|---|
| -w <dirname> | working directory |
| -f <filename> | file with feature table in tsv format: rows – features, columns – samples |
| --model <filename> | file with pre-trained classification model, obtained via fit or cv module |
Output files
wd_predict/predictions.tsv – tab-separated file with samples' names and predicted categories. As we can see by the results, categories for all samples were correctly predicted.
| sample | predicted category | true category |
|---|---|---|
| test_A | A | A |
| test_B | B | B |
| test_C | C | C |
Contact
Please report any problems directly to the GitHub issue tracker.
Also, you can send your feedback to abivanov@itmo.ru.
Authors:
- Software: Artem Ivanov (ITMO University) and Vladimir Popov (SPbSU)
- Testing: Artem Ivanov (ITMO University)
- Idea, supervisor: Vladimir Ulyantsev (ITMO University)
License
The MIT License (MIT)
See also
- MetaFast – a toolkit for comparison of metagenomic samples.
- MetaCherchant – a tool for analysing genomic environment within a metagenome.
- RECAST – a tool for sorting reads per their origin in metagenomic time series.