The limited version of this app can be accessed here: https://bixbeta.shinyapps.io/PCA-Explorer/
To run locally (strongly recommended), use the following instructions:
git clone this repository using git clone https://github.com/bixBeta/DESeq2-shiny.git
or using the green (Code/Clone) button on top of this page.
cd to DESeq2-shiny/PCA-Explorer
copy the path for the ui and server files by using pwd, we will need this path later.
Launch R Console on a Mac or PC and INSTALL the following packages:
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")
if (!requireNamespace("DESeq2", quietly = TRUE)) BiocManager::install("DESeq2")
if (!requireNamespace("dplyr", quietly = TRUE)) install.packages("dplyr")
if (!requireNamespace("plotly", quietly = TRUE)) install.packages("plotly")
if (!requireNamespace("shiny", quietly = TRUE)) BiocManager::install("shiny")
if (!requireNamespace("tibble", quietly = TRUE)) install.packages("tibble")
if (!requireNamespace("DT", quietly = TRUE)) install.packages("DT")
if (!requireNamespace("shinyjs", quietly = TRUE)) BiocManager::install("shinyjs")
Once all packages are installed:
From R Console type shiny::runApp('~/path/to/DESeq2-shiny/PCA-Explorer')# (paste the copied path here)
Alternatively, from a bash terminal type R -e shiny::runApp('~/path/to/DESeq2-shiny/PCA-Explorer')
Copy the ip address and paste it in a web browser to launch the app
Example CountMatrix and metadata target files are available in the PCA-Explorer/example folder.
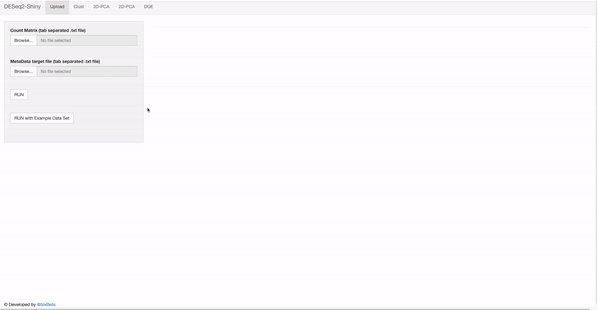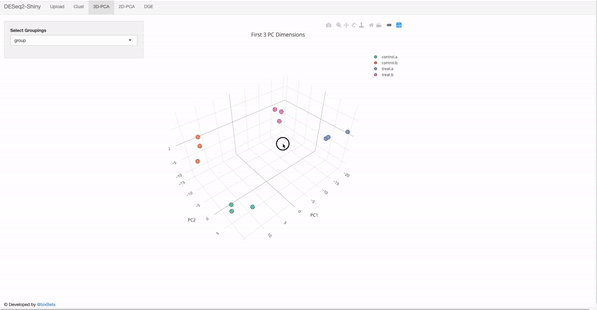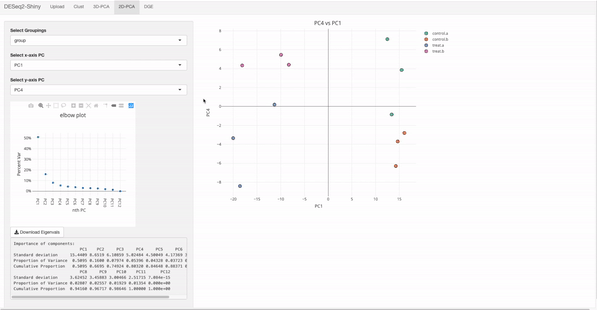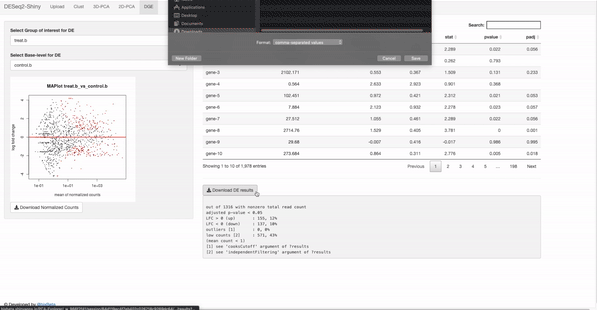
In the target file, label and group columns are mandatory.
In the label column, please refrain from using any special characters, only . is acceptable e.g. Sample.1
The label column fields must match the column names of the countMatrix.txt file.
e.g. Sample.1 label must have a corresponding Sample.1 counts column in the countMatrix.txt file
All labels are case-sensitive.