The TronFlow VCF postprocessing pipeline is part of a collection of computational workflows for tumor-normal pair somatic variant calling. These workflows are implemented in the Nextflow (Di Tommaso, 2017) framework.
This pipeline has several objectives:
- Variant filtering
- Variant normalization (BCFtools and vt)
- Technical annotations from different BAM files (VAFator)
- Functional annotations (SnpEff or BCFtools csq)
All of the previous steps are optional.
Run it from GitHub as follows:
nextflow run tron-bioinformatics/tronflow-vcf-postprocessing -r v2.1.0 -profile conda --input_vcfs input_vcfs --reference reference.fasta
Otherwise download the project and run as follows:
nextflow main.nf -profile conda --input_vcfs input_vcfs --reference reference.fasta
Find the help as follows:
$ nextflow run tron-bioinformatics/tronflow-vcf-postprocessing --help
TronFlow VCF postprocessing v${VERSION}
Usage:
nextflow run main.nf --input_vcfs input_vcfs --reference reference.fasta
Input:
* --input_vcf: the path to a single VCF to normalize (not compatible with --input_files)
* --input_vcfs: the path to a tab-separated values file containing in each row the sample name and path to the VCF file (not compatible with --input_vcf)
The input file does not have header!
Example input file:
sample1 /path/to/your/file.vcf
sample2 /path/to/your/file2.vcf
Optional input:
* --input_bams: a tab-separated values file containing in each row the sample name and path to a BAM file for VAFator annotations
The input file does not have header!
Example input file:
sample1 primary:/path/to/your/file.vcf
sample1 metastasis:/path/to/your/file2.vcf
* --input_purities: a tab-separated values file containing in each row the sample name and a purity value for VAFator annotations
The input file does not have header!
Example input file:
sample1 primary:0.5
sample1 metastasis:0.6
* --input_clonalities: a tab-separated values file containing in each row the sample name and a either a
genome-wide clonality value or a Bedgraph file with local clonality values for VAFator annotations
The input file does not have header!
Example input file:
sample1 primary:3
sample1 metastasis:/path/to/metastasis.local_clonalities.bed
* --reference: absolute path to the FASTA genome reference (indexes expected *.fai, *.dict) [required for normalization and for functional annotation with BCFtools]
* --gff: absolute path to a GFF gene annotations file [required for functional annotation with BCFtools, only Ensembl-like GFF files]
* --vcf-without-ad: indicate when the VCFs to normalize do not have the FORMAT/AD annotation
* --output: the folder where to publish output
* --skip_normalization: flag indicating to skip all normalization steps
* --skip_decompose_complex: flag indicating not to split complex variants (ie: MNVs and combinations of SNVs and indels)
* --filter: specify the filter to apply if any (e.g.: PASS), only variants with this value will be kept
* --input_bams: a tab-separated values file containing in each row the sample name, tumor and normal BAM files for annotation with Vafator
* --skip_multiallelic_filter: after VAFator annotations if any multiallelic variant is present (ie: two different
mutations in the same position) only the highest VAF variant is kept unless this flag is passed
* --snpeff_organism: the SnpEff organism name (eg: hg19, hg38, GRCh37.75, GRCh38.99)
* --snpeff_datadir: the SnpEff data folder where the reference genomes were previously downloaded. Required if --snpeff_organism is provided
* --snpeff_args: additional SnpEff arguments
* --snpeff_memory: for some samples SnpEff may require to use more memory (default: 3g)
* --mapping_quality: VAFator minimum mapping quality (default: 0)
* --base_call_quality: VAFator minimum base call quality (default: 0)
Output:
* Normalized VCF file
* Tab-separated values file with the absolute paths to the normalized VCF files, normalized_vcfs.txt
* Summary statistics before and after normalization
The table with VCF files expects two tab-separated columns without a header
| Patient name | VCF |
|---|---|
| patient_1 | /path/to/patient_1.vcf |
| patient_2 | /path/to/patient_2.vcf |
The optional table with BAM files expects two tab-separated columns without a header.
| Patient name | Sample name:BAM |
|---|---|
| patient_1 | primary_tumor:/path/to/sample_1.primary.bam |
| patient_1 | metastasis_tumor:/path/to/sample_1.metastasis.bam |
| patient_1 | normal:/path/to/sample_1.normal.bam |
| patient_2 | primary_tumor:/path/to/sample_1.primary_1.bam |
| patient_2 | primary_tumor:/path/to/sample_1.primary_2.bam |
| patient_2 | metastasis_tumor:/path/to/sample_1.metastasis.bam |
| patient_2 | normal:/path/to/sample_1.normal.bam |
Each patient can have any number of samples. Any sample can have any number of BAM files, annotations from the different BAM files of the same sample will be provided with suffixes _1, _2, etc. The aggregated vafator annotations on each sample will also be provided without a suffix.
The optional table with tumor purities expects two tab-separated columns without a header. The default purity is 1.0. Purity values are in the range 0.0 to 1.0. The purity values are used to adjust the expected VAF which is then used to calculate the power to detect a somatic mutation and the probability of an undetected somatic mutation.
| Patient name | Sample name:purity |
|---|---|
| patient_1 | primary_tumor:0.4 |
| patient_1 | metastasis_tumor:0.5 |
| patient_2 | primary_tumor:0.6 |
| patient_2 | metastasis_tumor:0.7 |
The optional table with local clonality values expects two tab-separated columns without a header. The local clonalities are provided in a Bedgraph file as described in VAFator documentation or as a genome-wide numeric parameter.
| Patient name | Sample name:local clonalities |
|---|---|
| patient_1 | primary_tumor:/path/to/patient_1.primary.local_clonalities.bed |
| patient_1 | metastasis_tumor:/path/to/patient_1.metastasis.local_clonalities.bed |
| patient_2 | primary_tumor:3 |
| patient_2 | metastasis_tumor:2 |
Only variants with the value in the column FILTER matching the value of parameter --filter are kept.
If this parameter is not used not variants are filtered out. Multiple values can be passed separated by commas without spaces.
For instance, --filter PASS,. will keep variant having FILTER=PASS or FILTER=., but remove all others.
No filter is applied if --filter is not passed.
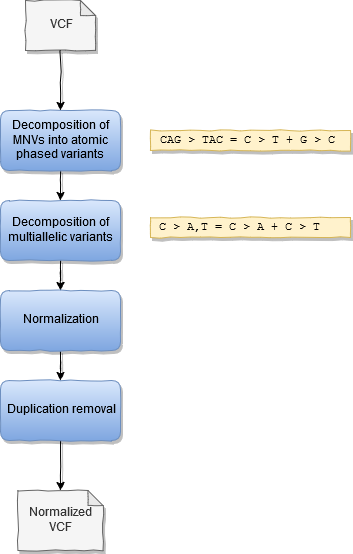
The normalization step aims to represent variants into the convened normal form as described in Tan 2015. The variant normalization is based on the implementation in vt (Tan, 2015) and bcftools (Danecek, 2021).
The normalization pipeline consists of the following steps:
- Decomposition of MNPs into atomic variants (ie: AC > TG is decomposed into two variants A>T and C>G) (optional).
- Decomposition of multiallelic variants into biallelic variants (ie: A > C,G is decomposed into two variants A > C and A > G)
- Trim redundant sequence and left align indels, indels in repetitive sequences can have multiple representations
- Remove duplicated variants
The output consists of:
- The normalized VCF
- Summary statistics before and after normalization
The parameter --reference pointing to the reference genome FASTA file is required for normalization.
The reference genome requires *.fai and *.dict indexes.
Normalization is not applied if the parameter --skip_normalization is passed.
Before:
chr1 13083 . AGA AGC . UNTRIMMED GT:AD 0:276,0 0/1:260,18
After:
chr1 13085 . A C . UNTRIMMED OLD_CLUMPED=chr1|13083|AGA|AGC GT:AD 0:276,276 0/1:260,260
NOTE: when a variant is changed during normalization the original variant is kept in the INFO field OLD_CLUMPED.
If an indel lies within a repetitive sequence it can be reported on different positions, the convention is to report the left most indel. This is what is called left alignment.
Before:
chr1 13141 . C AAAAAC . UNALIGNED GT:AD 0:80,1 0/1:76,14
chr1 13141 . CTGAGG G . UNALIGNED GT:AD 0:80,1 0/1:76,14
After:
chr1 13140 . C CAAAAA . UNALIGNED OLD_CLUMPED=chr1|13141|C|AAAAAC GT:AD 0:80,1 0/1:76,14
chr1 13140 . CCTGAG C . UNALIGNED OLD_CLUMPED=chr1|13141|CTGAGG|G GT:AD 0:80,1 0/1:76,14
NOTE: the rule of thumb to confirm if any given indel is left aligned, assuming that they are trimmed, is that the last base in both reference and alternate must differ.
Before:
chr1 13204 . C G,T . MULTIALLELIC GT:AD 1:229,1,1 0/1:196,24,1
After:
chr1 13204 . C G . MULTIALLELIC OLD_VARIANT=chr1|13204|C|G,|1 GT:AD 1:229,229 0/1:196,196
chr1 13204 . C T . MULTIALLELIC OLD_VARIANT=chr1|13204|C|G,|2 GT:AD 0:229,229 0/0:196,196
Note that the AD values are incorrectly set after the split, this seems to be a regression issue in bcftools v1.12 and has been reported here samtools/bcftools#1499.
Before:
chr1 13261 . GCTCCT CCCCCC . MNV GT:AD 0:41,0 0/1:35,3
After:
chr1 13261 . G C . MNV OLD_CLUMPED=chr1:13261:GCTCCT/CCCCCC GT:AD:PS 0:41,0:13261 0|1:35,3:13261
chr1 13263 . T C . MNV OLD_CLUMPED=chr1:13261:GCTCCT/CCCCCC GT:AD:PS 0:41,0:13261 0|1:35,3:13261
chr1 13266 . T C . MNV OLD_CLUMPED=chr1:13261:GCTCCT/CCCCCC GT:AD:PS 0:41,0:13261 0|1:35,3:13261
This behaviour is optional and can be disabled with --skip_decompose_complex. Beware, that the phase is maintained in
the fields FORMAT/GT and FORMAT/PS as described in the VCF specification section 1.4.2.
Before:
chr1 13321 . AGCCCT CGCC . MNV-INDEL GT:AD 0:229,1 0/1:196,24
After:
chr1 13321 . A C . MNV-INDEL OLD_CLUMPED=chr1:13321:AGCCCT/CGCC GT:AD:PS 0:229,1:13321 0|1:196,24:13321
chr1 13322 . GC G . MNV-INDEL OLD_CLUMPED=chr1:13321:AGCCCT/CGCC GT:AD:PS 0:229,1:13321 0|1:196,24:13321
chr1 13325 . CT C . MNV-INDEL OLD_CLUMPED=chr1:13321:AGCCCT/CGCC GT:AD:PS 0:229,1:13321 0|1:196,24:13321
Same as MNVs this behaviour can de disabled with --skip_decompose_complex.
The technical annotations provide an insight on the variant calling process by looking into the context of each variant within the pileup of a BAM file. When doing somatic variant calling it may be relevant to have technical annotations for the same variant in a patient from multiple BAM files. These annotations are provided by VAFator (https://github.com/TRON-Bioinformatics/vafator).
No technical annotations are performed if the parameter --input_bams is not passed.
The phase of the mutations can be inferred from the reads pileup.
This information is relevant to determine the impact of nearby mutations.
WhatsHap adds the phasing information when possible to the VCF following the VCF specification for that purpose.
Genotypes with phase information use the | instead of the / and 0|1 and 1|0 represent the two different phases.
A homozygous mutations in phase is represented as 1|1.
Furthermore, because phasing may be incomplete, to define the different phased blocks the INFO/PS annotation used.
All mutations belonging to the same phase block share the same PS number.
BAM files need to be provided to perform phasing with --input_bams and the phasing sample has to be determined with
--phasing_sample.
This approach has some limitations:
- We only support diploid genotyped VCF files. WhatsHap does support polyploid samples. But the particular case of somatic mutations is not supported.
- The sample name chosen to perform the phasing has to be the same across all VCFs.
Not to be mistaken with the patient name. Eg: only
normalsamples across all patients can be used for phasing
The functional annotations provide a biological context for every variant. Such as the overlapping genes or the effect of the variant in a protein. These annotations are provided by SnpEff (Cingolani, 2012) or by BCFtools csq (Danecek, 2017). Only one of the previous can be used.
The SnpEff available human annotations are:
- GRCh37.75
- GRCh38.99
- hg19
- hg19kg
- hg38
- hg38kg
Before running the functional annotations you will need to download the reference genome you need to use.
This can be done as follows: snpEff download -dataDir /your/snpeff/folder -v hg19
When running indicate the right reference genome like --snpeff_organism hg19.
If none is provided no SnpEff annotations will be provided.
Provide the snpEff folder with --snpeff_datadir
To provide any additional SnpEff arguments use --snpeff_args such as
--snpeff_args "-noStats -no-downstream -no-upstream -no-intergenic -no-intron -onlyProtein -hgvs1LetterAa -noShiftHgvs",
otherwise defaults will be used.
No SnpEff functional annotations are performed if the parameters --snpeff_organism and --snpeff_datadir are not passed.
BCFtools does not require any previous preparation. It expects two parameters:
--reference: absolute path to the FASTA reference genome--gff: absolute path to the Ensembl-like GFF annotations (ie: Gencode GFF files do not work samtools/bcftools#1078)
Importantly, BCFtools does use the available phasing information to evaluate all mutations affecting any given transcript together.
- Adrian Tan, Gonçalo R. Abecasis and Hyun Min Kang. Unified Representation of Genetic Variants. Bioinformatics (2015) 31(13): 2202-2204](http://bioinformatics.oxfordjournals.org/content/31/13/2202) and uses bcftools [Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (Oxford, England), 27(21), 2987–2993. 10.1093/bioinformatics/btr509
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021 Feb 16;10(2):giab008. doi: 10.1093/gigascience/giab008. PMID: 33590861; PMCID: PMC7931819.
- Di Tommaso, P., Chatzou, M., Floden, E. W., Barja, P. P., Palumbo, E., & Notredame, C. (2017). Nextflow enables reproducible computational workflows. Nature Biotechnology, 35(4), 316–319. 10.1038/nbt.3820
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3.". Fly (Austin). 2012 Apr-Jun;6(2):80-92. PMID: 22728672
- Danecek, P., & McCarthy, S. A. (2017). BCFtools/csq: haplotype-aware variant consequences. Bioinformatics, 33(13), 2037–2039. https://doi.org/10.1093/bioinformatics/btx100