This pipeline has moved!
This pipeline has been moved to the new nf-core project. You can now find it here:
https://github.com/nf-core/neutronstar
If you have any problems with the pipeline, please create an issue at the above repository instead.
To find out more about nf-core, visit http://nf-co.re/
This repository will be archived to maintain the released versions for future reruns, in the spirit of full reproducibility.
If you have any questions, please get in touch: support@ngisweden.se
// Phil Ewels, 2018-10-05
Table of Contents
Introduction
NGI-NeutronStar is a bioinformatics best-practice analysis pipeline used for de-novo assembly and quality-control of 10x Genomics Chromium data. It's developed and used at the National Genomics Infastructure at SciLifeLab Stockholm, Sweden. The pipeline uses Nextflow, a bioinformatics workflow tool.
Disclaimer
This software is in no way affiliated with nor endorsed by 10x Genomics.
Installation
Nextflow runs on most POSIX systems (Linux, Mac OSX etc). It can be installed by running the following commands:
# Make sure that Java v7+ is installed:
java -version
# Install Nextflow
curl -fsSL get.nextflow.io | bash
# Add Nextflow binary to your PATH:
mv nextflow ~/bin
# OR system-wide installation:
# sudo mv nextflow /usr/local/binYou need NextFlow version >= 0.25 to run this pipeline.
While it is possible to run the pipeline by having nextflow fetch it directly from GitHub, e.g nextflow run SciLifeLab/NGI-NeutronStar, depending on your system you will most likely have to download it (and configure it):
get https://github.com/SciLifeLab/NGI-NeutronStar/archive/master.zip
unzip master.zip -d /my-pipelines/
cd /my_data/
nextflow run /my-pipelines/NGI-NeutronStar-masterSingularity
If running the pipeline using the Singularity configurations (see below), Nextflow will automatically fetch the image from DockerHub. However if your compute environment does not have access to the internet you can build the image elsewhere and run the pipeline using:
# Build image
singularity pull --name "ngi-neutronstar.simg" docker://remiolsen/ngi-neutronstar
# After uploading it to your_hpc:/singularity_images/
nextflow run -with-singularity /singularity_images/ngi-neutronstar.simg /my-pipelines/NGI-NeutronStar-masterBusco data
By default NGI-NeutronStar will look for the BUSCO lineage datasets in the data folder, e.g. /my-pipelines/NGI-NeutronStar-master/data/. However if you have these datasets installed any other path it is possible to specify this using the option --BUSCOfolder /path/to/lineage_sets/. Included with the pipeline is a script to download BUSCO data, in /my-pipelines/NGI-NeutronStar-master/data/busco_data.py
# Example downloading a minimal, but broad set of lineages
cd /my-pipelines/NGI-NeutronStar-master/data/
# To list the datasets
# Category minimal contains:
# - bacteria_odb9
# - eukaryota_odb9
# - metazoa_odb9
# - protists_ensembl
# - embryophyta_odb9
# - fungi_odb9
python busco_data.py list minimal
# To download them
python busco_data.py download minimalUsage instructions
It is recommended that you start the pipeline inside a unix screen (or alternatively tmux).
Single assembly
To assemble a single sample, the pipeline can be started using the following command:
nextflow run \\
-profile nextflow_profile \\
/path/to/NGI-NeutronStar/main.nf \\
[Supernova options] \\
(--clusterOptions)nextflow_profileis one of the environments that are defined in the file nextflow.config[Supernova options]are the following options that are following supernova options (use the commandsupernova run --helpfor a more detailed description or alternatively read the documentation available by 10X Genomics)--fastqsrequired--idrequired--sample--lanes--indices--bcfrac--maxreads
--clusterOptionsare the options to feed to the HPC job manager. For instance for SLURM--clusterOptions="-A project -C node-type"--genomesizerequired The estimated size of the genome(s) to be assembled. This is mainly used by Quast to compute NGxx statstics, e.g. N50 statistics bound by this value and not the assembly size.
Multiple assemblies
NGI-NeutronStar also supports adding the above parameters in a .yaml file. This way you can run several assemblies in parallel. The following example file (sample_config.yaml) will run two assemblies of the test data included in the Supernova installation, one using the default parameters, and one using barcode downsampling:
genomesize: 1000000
samples:
- id: testrun
fastqs: /sw/apps/bioinfo/Chromium/supernova/1.1.4/assembly-tiny-fastq/1.0.0/
- id: testrun_bc05
fastqs: /sw/apps/bioinfo/Chromium/supernova/1.1.4/assembly-tiny-fastq/1.0.0/
maxreads: 500000000
bcfrac: 0.5Run nextflow using nextflow run -profile -params-file sample_config.yaml /path/to/NGI-NeutronStar/main.nf (--clusterOptions)
Advanced usage
If not specifying the option -profile it will use a default one that is suitable to testing the pipeline on a typical laptop computer (using the test dataset included with the Supernova package). In a high-performance computing environment (and with real data) you should specify one of the hpc profiles. For instance for a compute cluster with the Slurm job scheduler and Singularity version >= 2.4 installed, hpc_singularity_slurm.
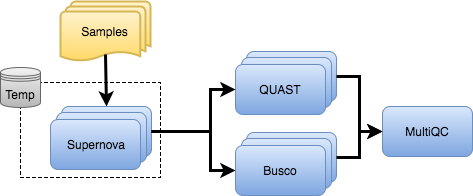
Pipeline overview
Credits
These scripts were written for use at the National Genomics Infrastructure at SciLifeLab in Stockholm, Sweden. Written by Remi-Andre Olsen (@remiolsen).