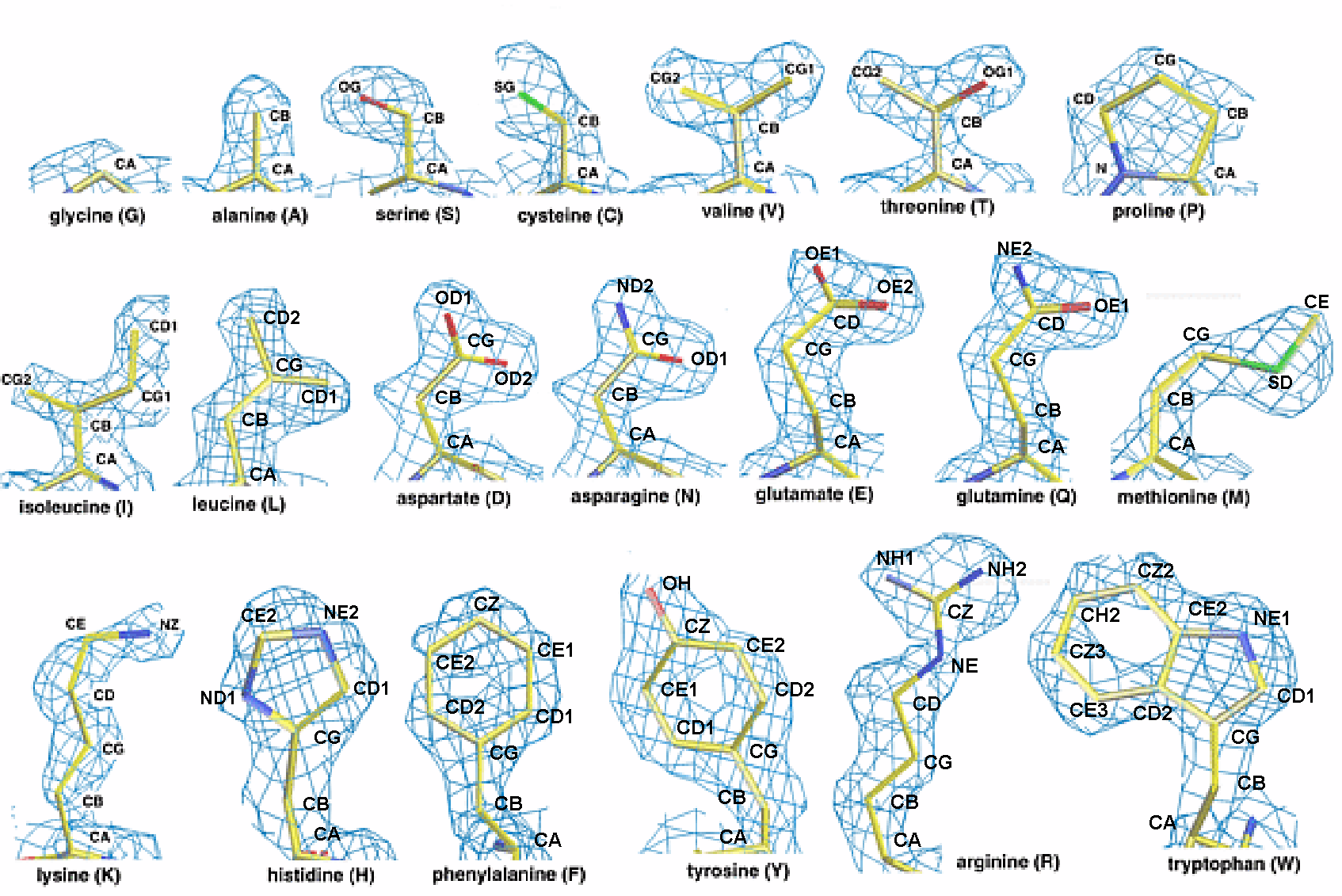
http://melroleandro.github.io/Protein-periodicity/
This is the base for a project on packing charged multibody systems with implicit solvation. This scripts are used for the extraction of statistical dada from large samples of crystal structures, selected from the PDB. The generated statistics are used on two type of libraries for multibody dynamics: a library with statistical potentials for pairs of dihedral angles in the main chain local conformation, and a rotamer libraries. This libraries are defined as a set of discrete probability distribution, and used to generat internal forces multibody systems.
-
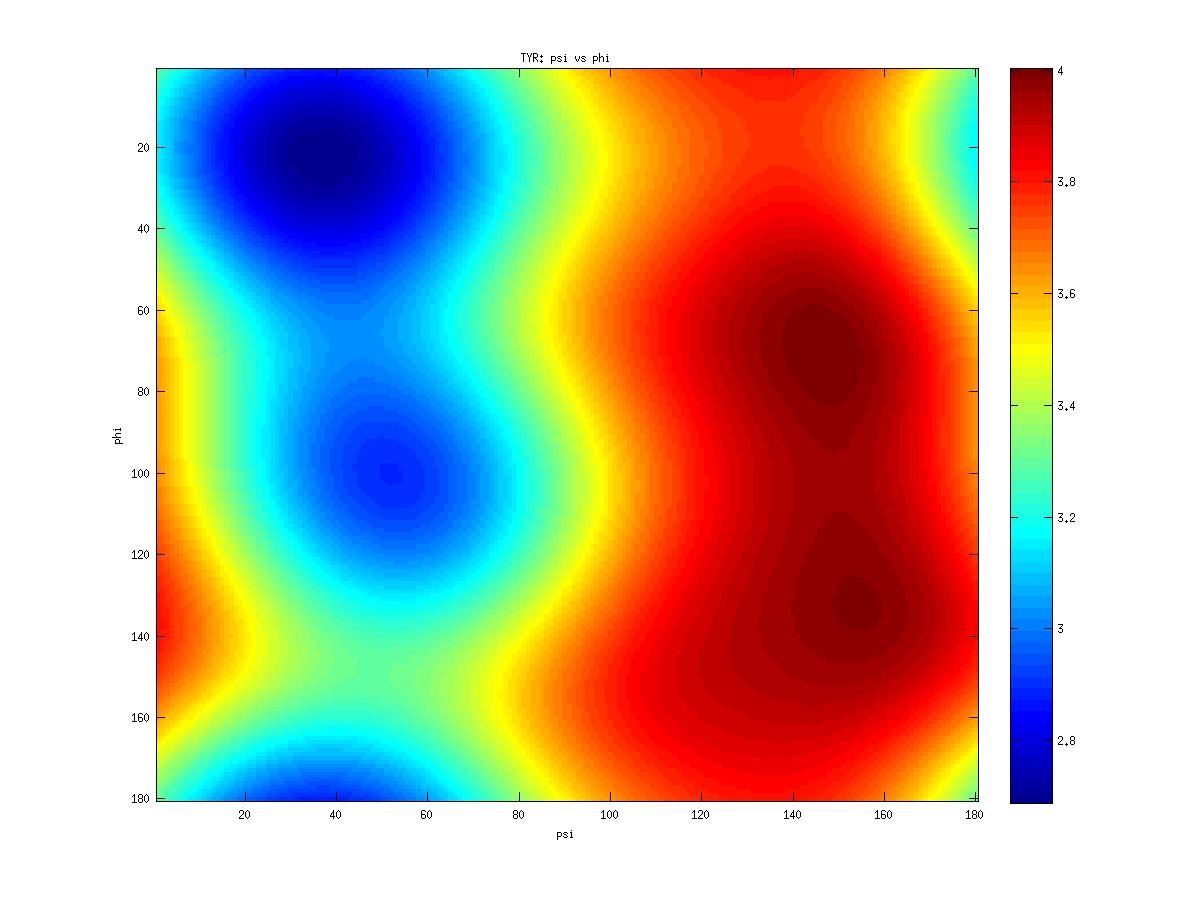
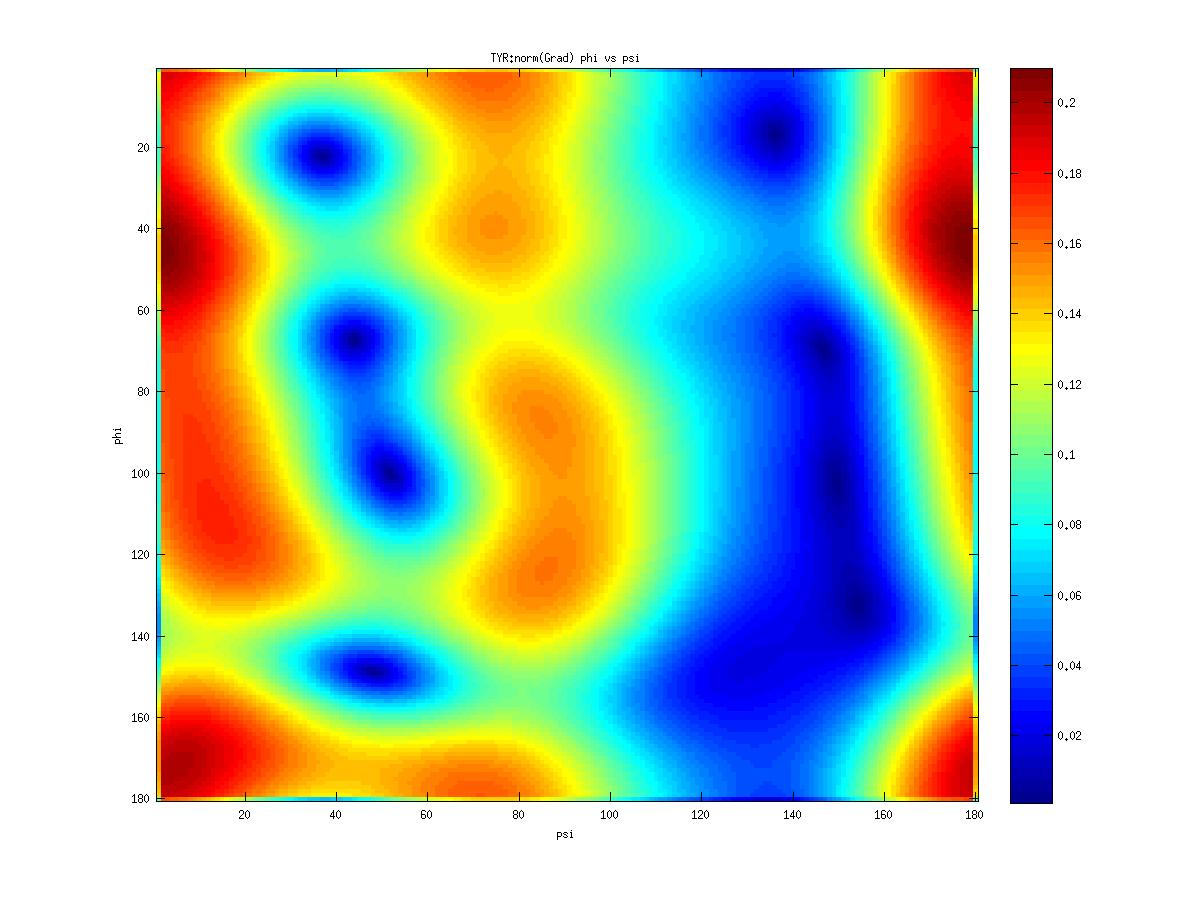
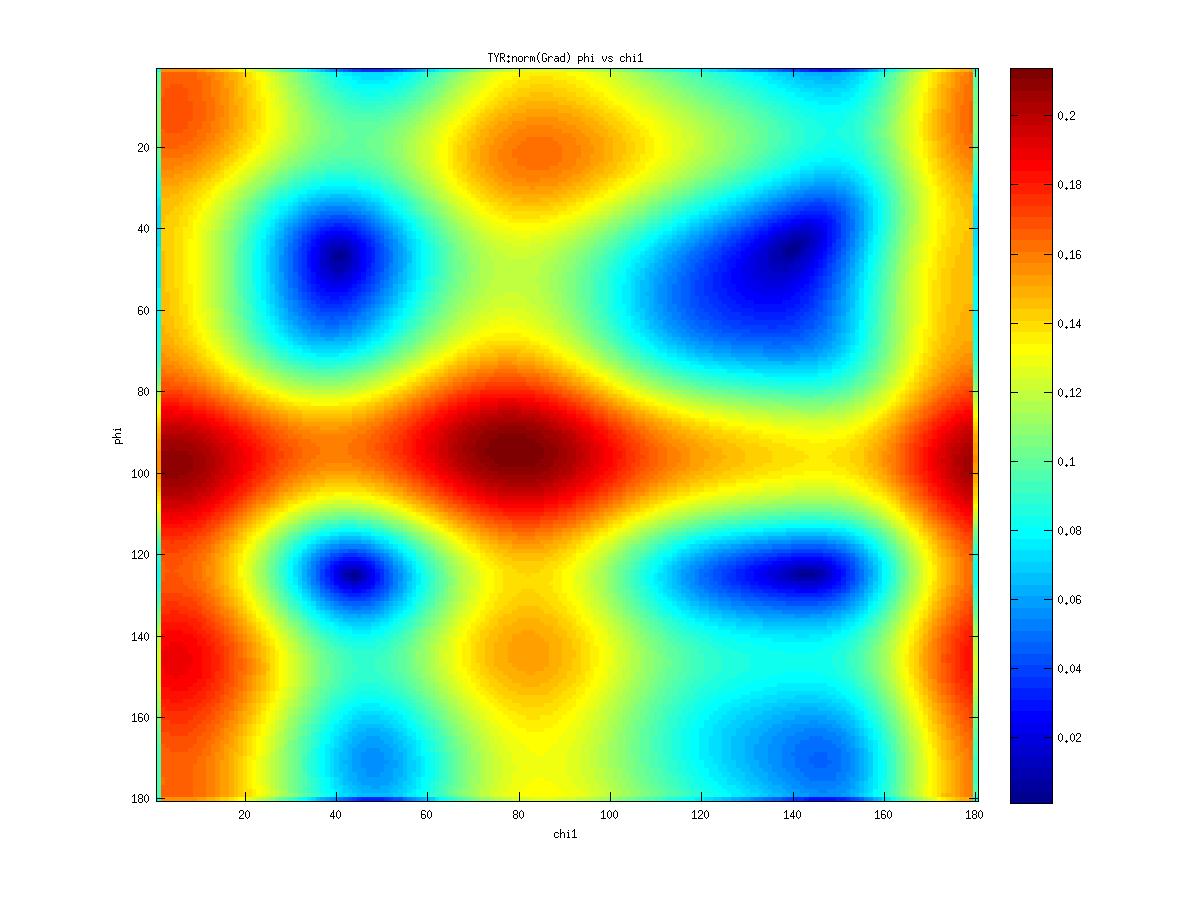
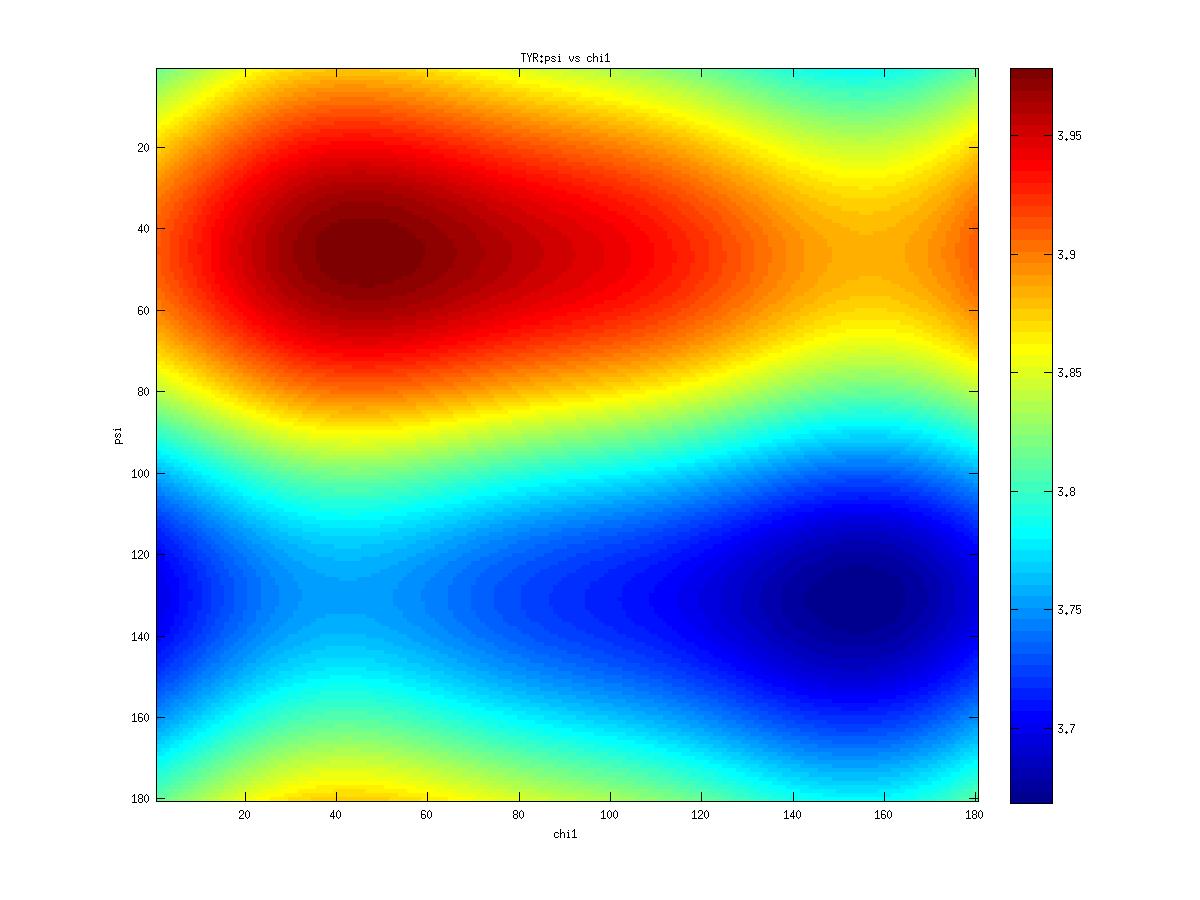
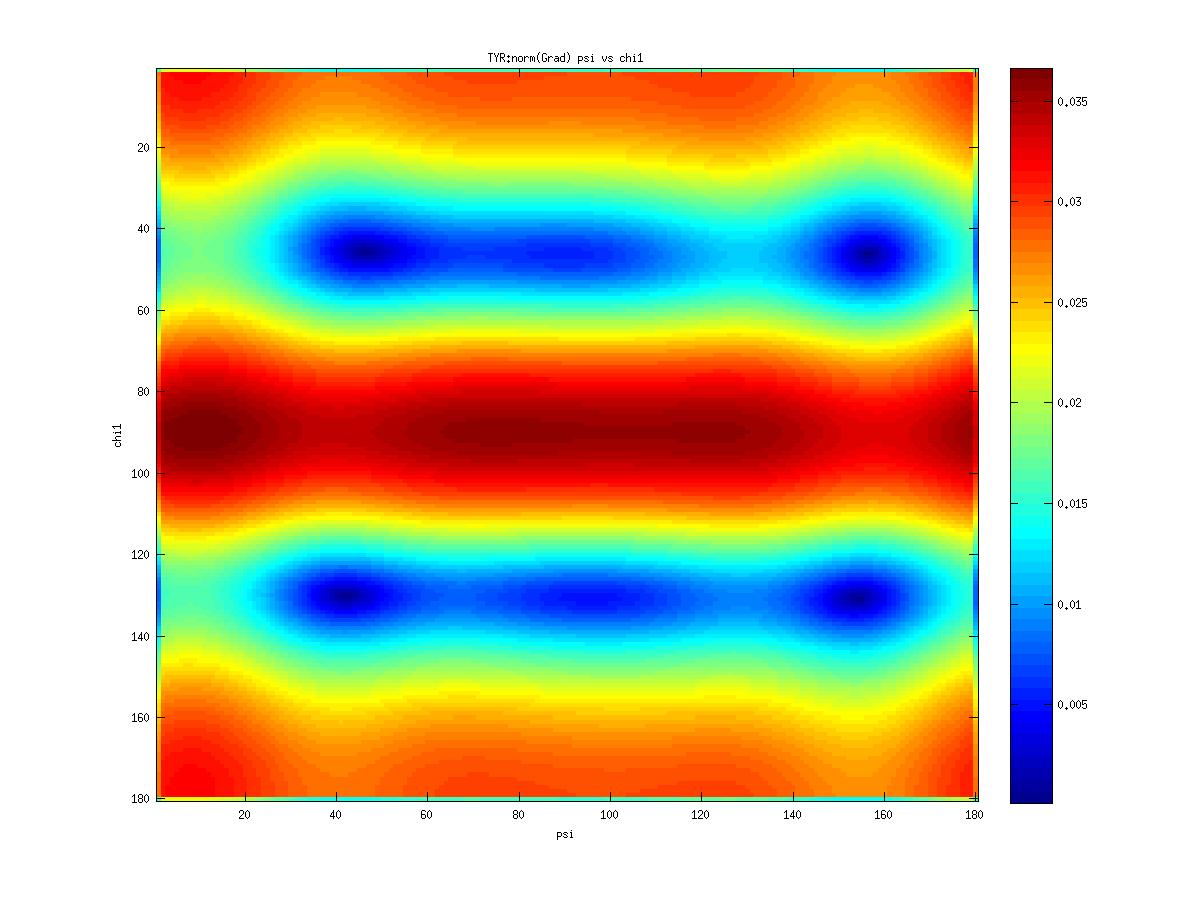
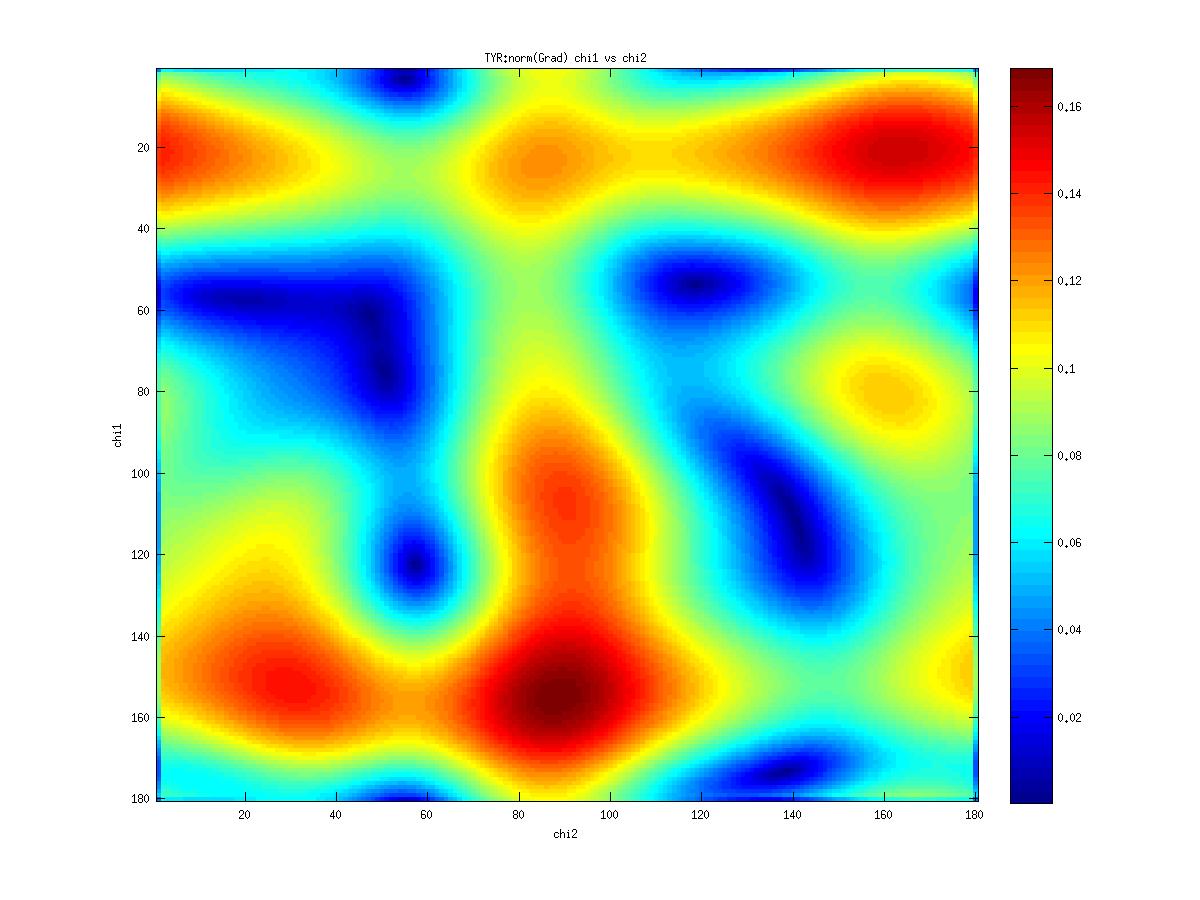
Examples of statistical potentials generated from two dihedral angles on a protein main chain:
-
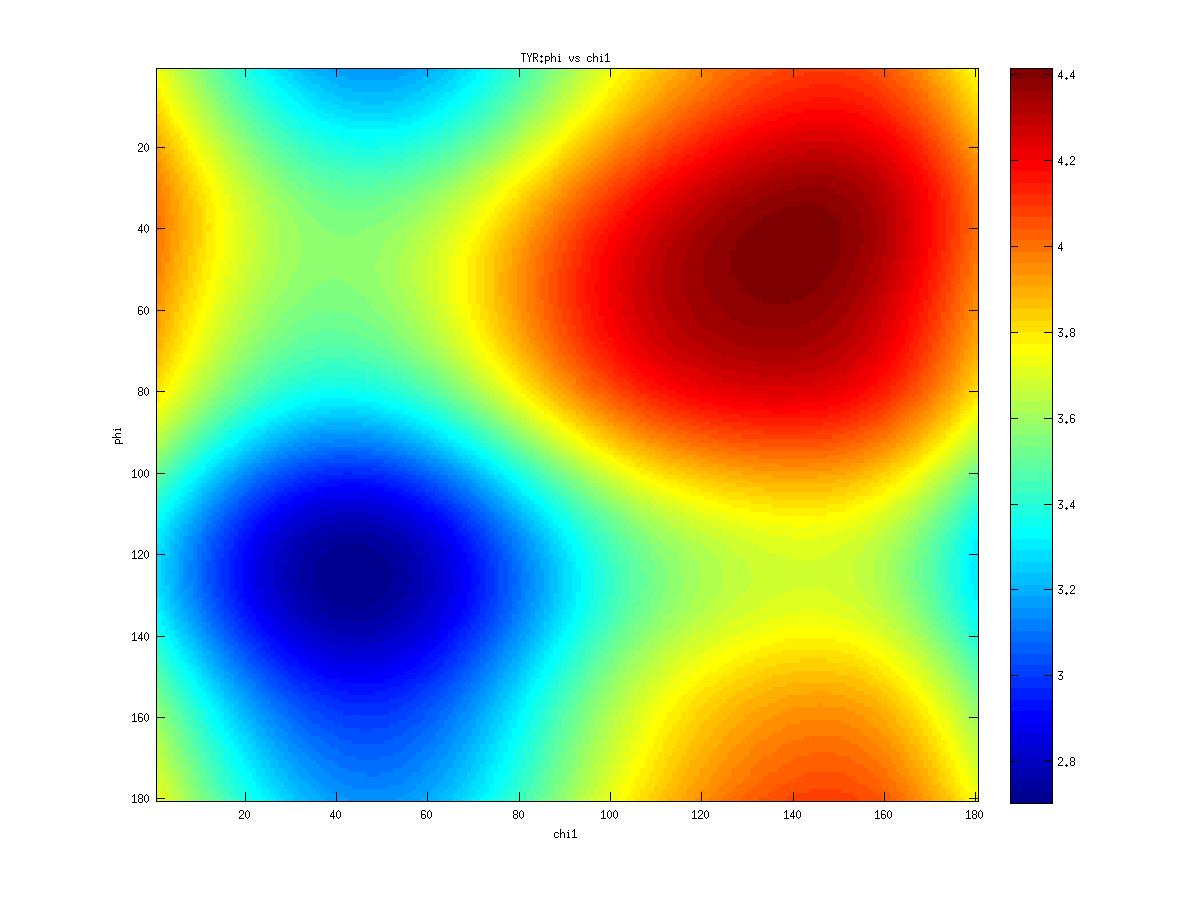
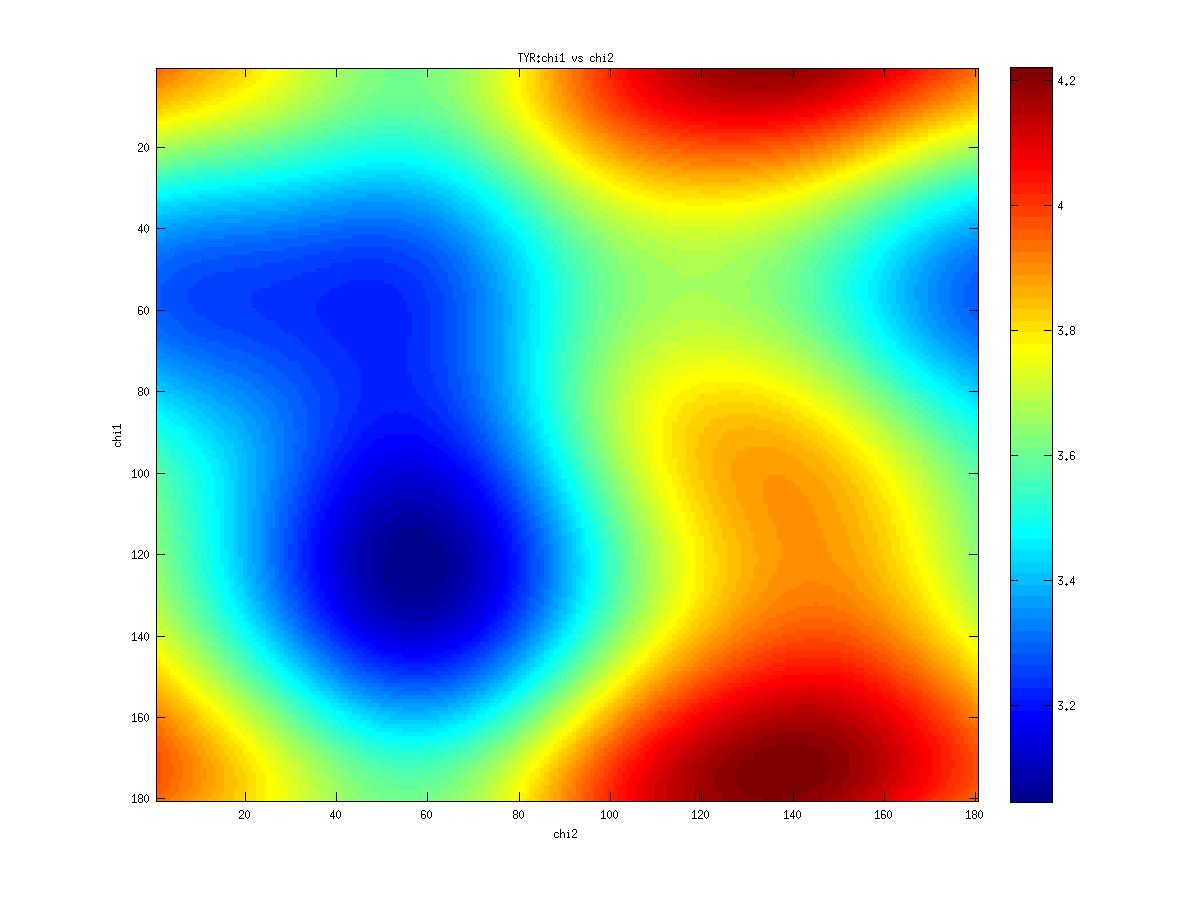
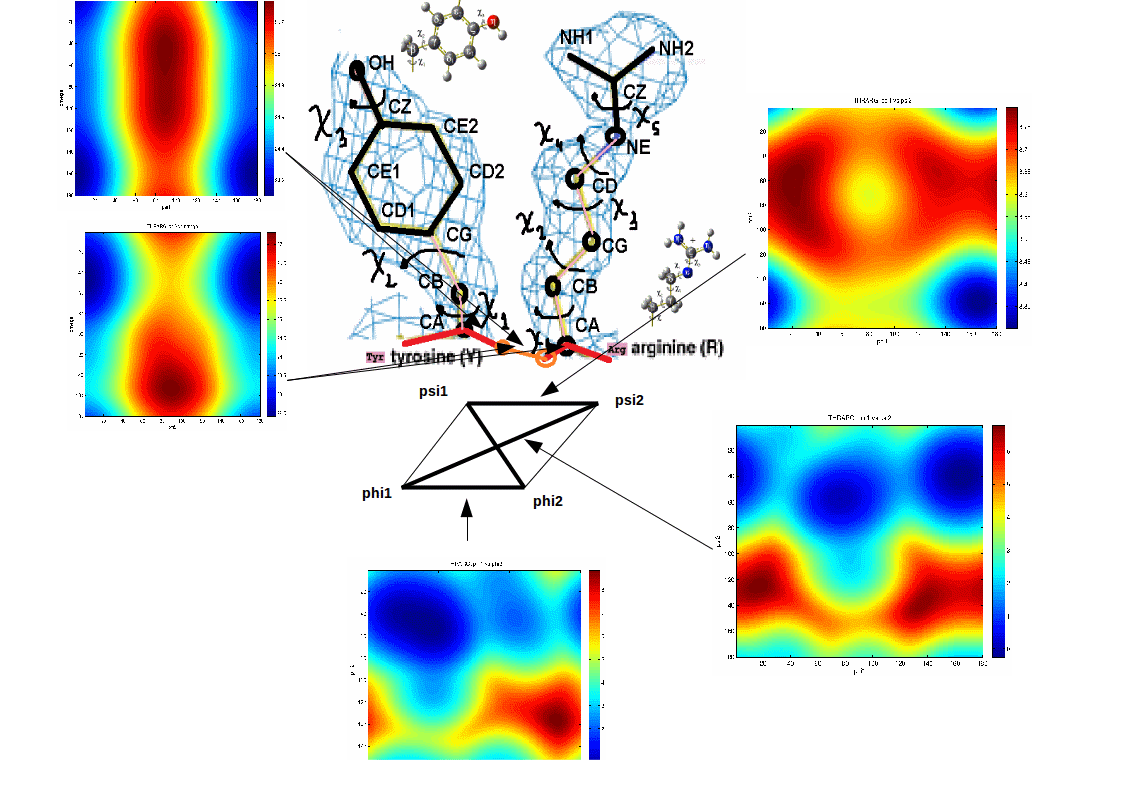
Examples of statistical potentials generated between dihedral angles and rotamers in an amino acid:
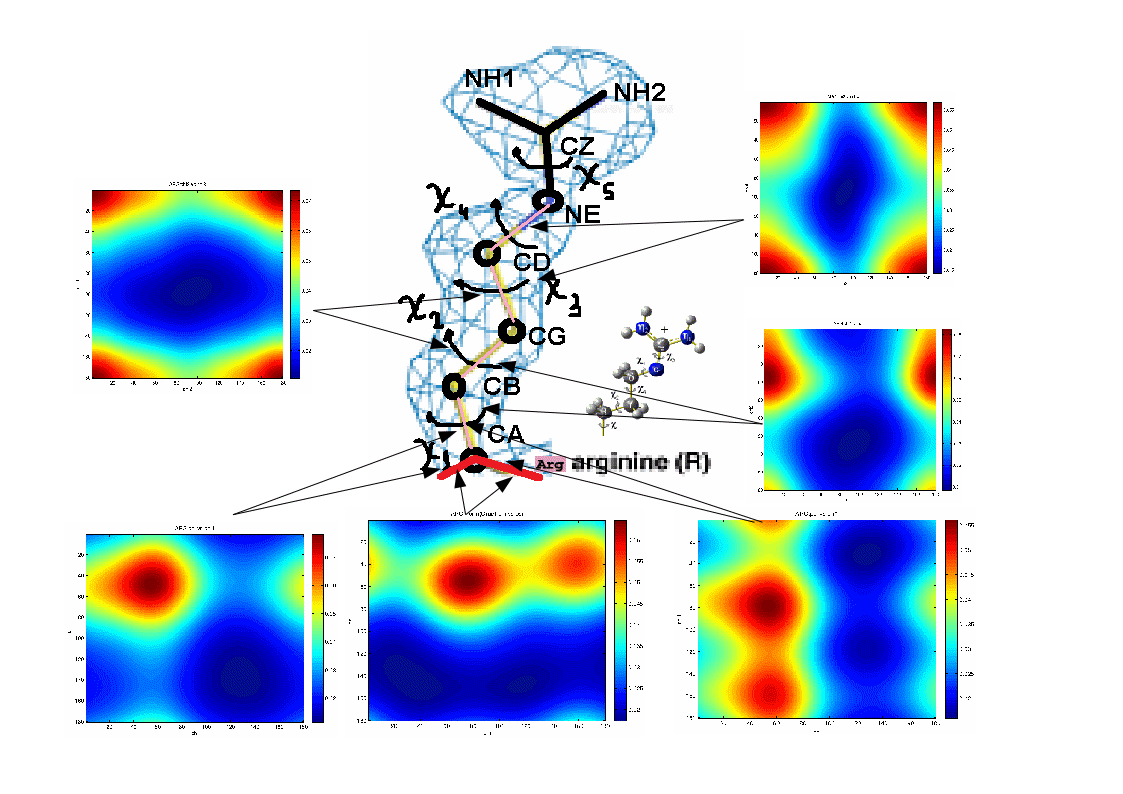
For more information: http://melroleandro.github.io/Protein-periodicity/
SAMPLE SIZE: 1615
| B-factor | |
|---|---|
| Mean | 18.34 |
| Median | 15.38 |
| Variance | 144.59 |
| Standard deviation | 12.02 |
| Skewness | 2.73 |
| Kurtosis | 15.87 |
| C-CA | N-CA | peptid plane | |
|---|---|---|---|
| Mean | 2.08 | 1.79 | 1.60 |
| Median | 1.53 | 1.46 | 1.33 |
| Variance | 1.50 | 0.77 | 0.54 |
| Standard deviation | 1.23 | 0.88 | 0.73 |
| Skewness | 2.00 | 2.85 | 3.07 |
| Kurtosis | 5.47 | 10.28 | 12.80 |
| corrcoef | 1.00 | 0.67 | 0.22 |
| corrcoef | 0.67 | 1.00 | 0.15 |
| corrcoef | 0.22 | 0.15 | 1.00 |
| PHI | PSI | |
|---|---|---|
| Mean | -89.45 | 111.99 |
| Median | -88.90 | 122.94 |
| R Length | 0.48 | 0.14 |
| Variance | 0.52 | 0.86 |
| Standard deviation | 1.02 | 1.31 |
| Standard deviation 0 | 1.21 | 1.97 |
| Skewness | -0.10 | 0.48 |
| Kurtosis | 0.34 | 0.18 |
Tests for Uniformity
- Rayleigh Test, P = 0.00 0.00
- Omnibus Test, P = 0.00 0.00
- Rao Spacing Test, P = 0.50 0.50
- V Test (r = 0), P = 0.40 1.00
Circular-Circular Association
- Circ-circ corr phi-psi coeff/pval: -0.22 0.000
- Circ-line corr phi-C-CA coeff/pval: 0.65 0.000
- Circ-line corr phi-N-CA coeff/pval: 0.43 0.000
- Circ-line corr psi-C-CA coeff/pval: 0.32 0.000
- Circ-line corr psi-N-CA coeff/pval: 0.21 0.000
| CHI_1 | |
|---|---|
| Mean resultant vector | 108.13 |
| Median | 108.13 |
| R Length | 0.05 |
| Variance | 0.95 |
| Standard deviation | 1.38 |
| Standard deviation 0 | 2.41 |
| Skewness | -0.00 |
| Kurtosis | 0.00 |
- Rayleigh Test, P = 0.01
- Omnibus Test, P = 0.05
- Rao Spacing Test, P = 0.50
- Circ-circ corr phi-chi1 coeff/pval: 0.00 0.850
- Circ-circ corr psi-chi1 coeff/pval: 0.04 0.141
| CHI_2 | |
|---|---|
| Mean resultant vector | -86.02 |
| Median | -86.02 |
| R Length | 0.38 |
| Variance | 0.62 |
| Standard deviation | 1.11 |
| Standard deviation 0 | 1.38 |
| Skewness | 0.27 |
| Kurtosis | 0.19 |
- Rayleigh Test, P = 0.00
- Omnibus Test, P = 0.00
- Rao Spacing Test, P = 0.50
- Circ-circ corr phi-chi2 coeff/pval: -0.16 0.000
- Circ-circ corr psi-chi2 coeff/pval: 0.06 0.011
- Circ-circ corr chi1-chi2 coeff/pval: 0.01 0.776
| CHI_3 | |
|---|---|
| Mean resultant vector | 11.78 |
| Median | 11.78 |
| R Length | 0.24 |
| Variance | 0.76 |
| Standard deviation | 1.23 |
| Standard deviation 0 | 1.68 |
| Skewness | 0.25 |
| Kurtosis | -0.63 |
- Rayleigh Test, P = 0.00
- Omnibus Test, P = 0.00
- Rao Spacing Test, P = 0.50
- Circ-circ corr phi-chi3 coeff/pval: 0.00 0.983
- Circ-circ corr psi-chi3 coeff/pval: -0.01 0.788
- Circ-circ corr chi1-chi3 coeff/pval: 0.00 0.960
- Circ-circ corr chi2-chi3 coeff/pval: -0.36 0.000