Shiny app for creating Venn diagrams of SARS-CoV-2 variant/lineage mutations.
Clone the repo from github.
git clone https://github.com/MDU-PHL/covid-venn.git && cd covid-venn.git
Install dependencies.
install.packages('shiny', 'VennDiagram', 'RColorBrewer')
Start the shiny server.
shiny::runApp()Shiny server can also be started from bash
Rscript -e "shiny::runApp()"Navigate to the URL printed in the console.
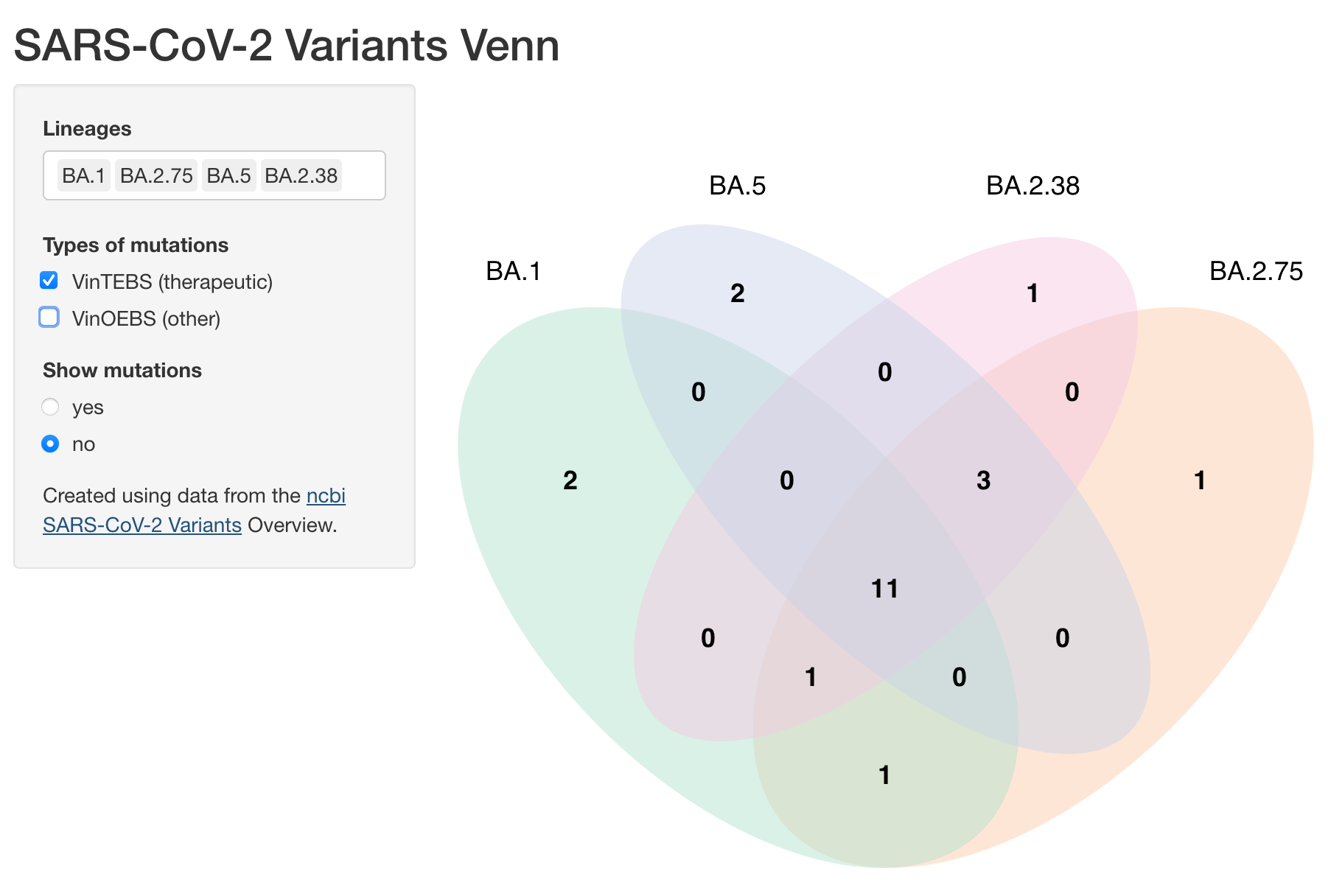
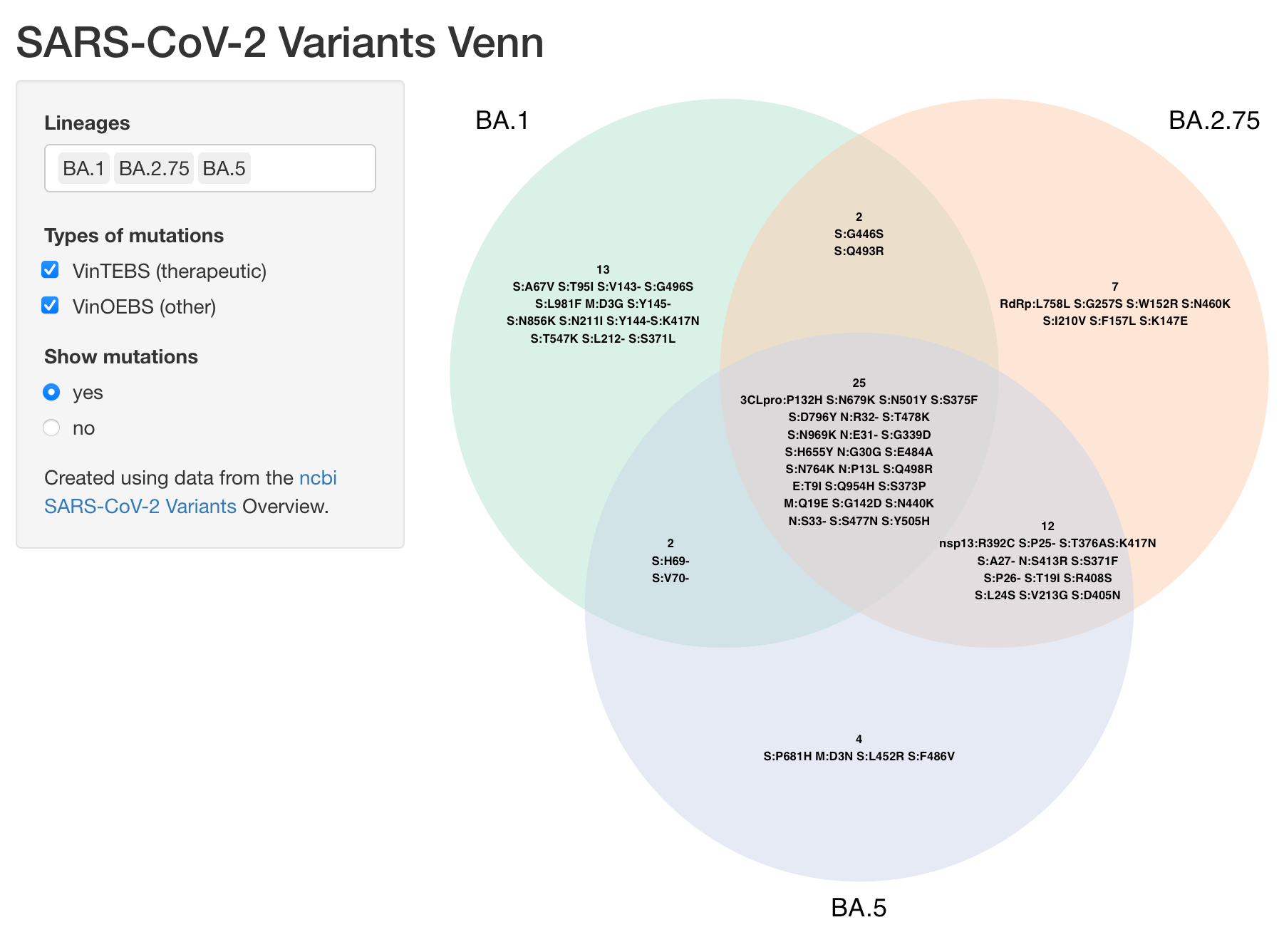
By default 3 lineages are selected. You can modify this selection using the Lineages select input. Up to 4 lineages can be compared.
There are three types of mutations that can be selected: Defining, VinTEBS (Variations in therapeutic epitopes or binding sites), and VinOEBS (Variations in other epitopes or binding sites).
Mutation labels can be shown using the Show mutations radio buttons.
Covid-venn uses data from the ncbi and downloads the latest version of the database each time it runs. If the database is unavailable covid-venn will default to using a local copy of the database (which may be out of date).