Codes for the paper : Predicting genotype environmental range from genome–environment associations
Stéphanie Manel, Marco Andrello, Karine Henry, Daphné Verdelet, Aude Darracq, Pierre‐Edouard Guerin, Bruno Desprez, Pierre Devaux
Molecular Ecology, may 2017
This repository contains all the scripts to calculate metrics (nucleotide diversity, Tajima's D...) on the beets genome from SNP data.
These metrics were necessary to validate our approach to predict the environmental range of species genotypes from the genetic markers significantly associated with those environmental variables in an independent set of individuals.
We applied this approach to predict aridity in a database constituted of 950 individuals of wild beets and 299 individuals of cultivated beets genotyped at 14,409 random single nucleotide polymorphisms (SNPs).
This study was funded by the French Government, under the management of the Research National Agency (ANR‐11‐BTBR‐0007) through the AKER programme in collaboration with Florimond Desprez company.
Prerequisites
Softwares
You must install the following softwares :
- VCFTOOLS
- TABIX & BGZIP
- R Version 3.2.3
R-packageggplot2R-packagePopGenome
- Python 2.7.12
Data Files
The included data files are :
- Positions.14409.txt: List of ID|position|scaffold|chromosome of 14409 SNPs.
- Data.950.sauvages.txt: List of names of the 950 indivuals of interest.
- NoPool.14409.csv: Table of genotypes of all the indivuals for the 14409 SNPs.
- Noms.marqueurs.LFMM.gINLAnd.csv: list of SNP IDs and results from Ginland & LFMM methods
- bon_exemple.vcf: Template of VCF format file used to create VCF files.
Code sources
scripts used to calculate statistics on the genome from SNP data
BASH scripts
- vcf4PopGenome_protocole.sh : Creates VCF.GZ files using TABIX, BGZIP and VCFTOOLS for each chromosome into mes_vcf/. Uncompressed VCF files will be saved into a new mes_vcf_save/.
- fabrique_outlier.sh: Generates a list of outliers SNPs positions for each chromosome into mes_outliers/.
- add_ID_to_tables.sh : Creates tables with ID of SNPs into tables/avec_id folder from tables/ results.
Python scripts
- get_col.py: Select columns of a CSV file according to a list of colunm's names.
- convert_data2vcf.py : Creates VCF files for each chromosome of each SNP with genotype of each indivuals.
- get_id_snp.py: get position|chromosome information in a table of SNP and find his ID in a VCF file.
R scripts
- fabrique_outlier.R: From Noms.marqueurs.LFMM.gINLAnd.csv, it provides a list of outliers SNPs IDs nom_84_outliers.txt
- generate_fig_tab.R : Generates sliding windows genome statistics into figures/ and tables/ folders using mes_vcfs/ and mes_outliers/ data.
- analysis_tables.R : Basic statistical analysis on
tables/avec_id/all_stats_propre.csv
Workflow
Calculate metrics (nucleotide diversity, Tajima's D...) on outliers and non-outliers SNPs and analysis
1. Creates VCF files of all the SNP and selected individuals
- Run vcf4PopGenome_protocole.sh to create VCF.GZ files into mes_vcf/ and VCF files into mes_vcf_save/, using data files and bon_exemple.vcf
bash vcf4PopGenome_protocole.sh
2. Get outliers SNPs and their positions
- Run fabrique_outlier.R to create nom_84_outliers.txt, the list of outliers SNPs IDs
- Run fabrique_outlier.sh to create a list of outliers SNPs positions for each chromosome into mes_outliers/
bash fabrique_outlier.sh
3. Generates figures and table of genome sliding 20Kbp-windows metrics
- Run generate_fig_tab.R to generate plot .PDF figures into figures/ folder and .CSV tables into tables/ for each chromosome
Rscript generate_fig_tab.R
4. Analyse statistics on tables
- Run add_ID_to_tables.sh to generate a merged table tables/avec_id/all_stats_propre.csv with SNPs IDs and statistics of all the chromosomes from tables/ to tables/avec_id/
- Run analysis_tables.R to have outliers SNP and non-outliers SNP statistics, then do some basic statistical tests.
bash add_ID_to_tables.sh
Rscript analysis_tables.R
Results
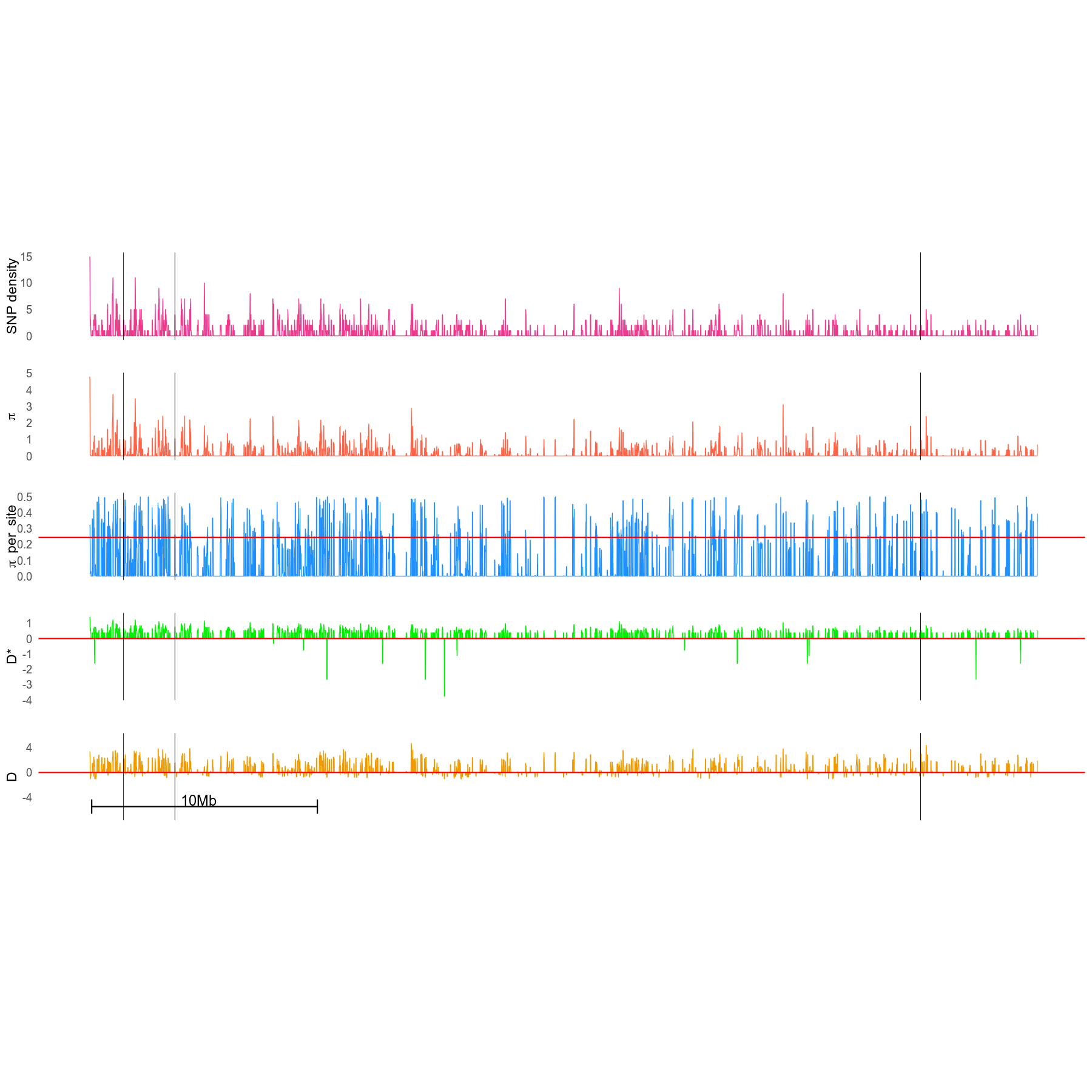
Beets genome chromosome 1 sequence: SNPs-metrics and significantly associated with aridity SNPs positions
Every 5 Kbp nucleotide steps, we calculated following metrics on a 20Kbp windows onto the genome :
- SNP density: number of SNPs
- pi : nucleotide diversity
- D: Tajima D
- D*: Fu and Li D
Positions of genetic markers SNP significantly associated with aridity environmental variable are indicated by a black dot