GenStore: A High-Performance and Energy-Efficient In-Storage Computing System for Genome Sequence Analysis
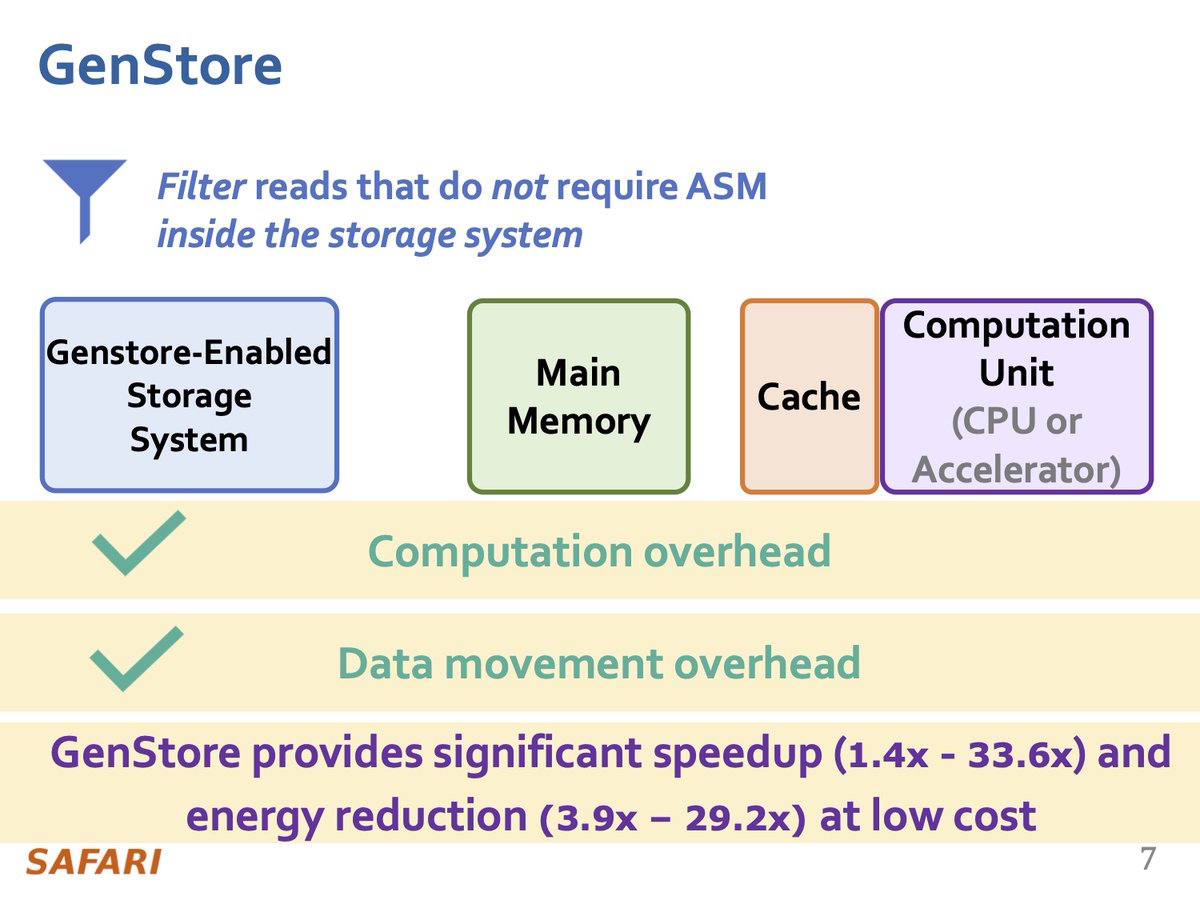
GenStore is the first in-storage processing system designed for genome sequence analysis that greatly reduces both data movement and computational overheads of genome sequence analysis by exploiting low-cost and accurate in-storage filters. GenStore leverages hardware/software co-design to address the challenges of in-storage processing, supporting reads with 1) different properties such as read lengths and error rates, which highly depend on the sequencing technology, and 2) different degrees of genetic variation compared to the reference genome, which highly depends on the genomes that are being compared.
Watch our full talk video (slides) and lightning talk video (slides) about GenStore!
If you find this repo useful, please cite the following paper:
Nika Mansouri Ghiasi, Jisung Park, Harun Mustafa, Jeremie Kim, Ataberk Olgun, Arvid Gollwitzer, Damla Senol Cali, Can Firtina, Haiyu Mao, Nour Almadhoun Alserr, Rachata Ausavarungnirun, Nandita Vijaykumar, Mohammed Alser, and Onur Mutlu, "GenStore: A High-Performance and Energy-Efficient In-Storage Computing System for Genome Sequence Analysis" Proceedings of the 27th International Conference on Architectural Support for Programming Languages and Operating Systems (ASPLOS), 2022
@inproceedings{mansouri2022genstore,
title={GenStore: a high-performance in-storage processing system for genome sequence analysis},
author={Mansouri Ghiasi, Nika and Park, Jisung and Mustafa, Harun and Kim, Jeremie and Olgun, Ataberk and Gollwitzer, Arvid and Senol Cali, Damla and Firtina, Can and Mao, Haiyu and Almadhoun Alserr, Nour and others},
booktitle={Proceedings of the 27th ACM International Conference on Architectural Support for Programming Languages and Operating Systems},
year={2022}
}- What is GenStore?
- Citation
- Prerequisites
- Preparing Input Data
- Baseline Software Exact Match Filter
- Software GenStore
- Hardware GenStore
- Contact
The infrastructure has been tested with the following system configuration:
- g++ v11.1.0
- Python v3.6
Prerequisites specific to each experiment are listed in their respective subsections.
The read sets used in the paper can be obtained by searching the read set eccession IDs provided in the paper in the European Bioinformatics Institute ftp.
We use mason_simulator (part of the SeqAn package) to simulate short reads of varying degree of genetic distance from the reference genome.
cd input-generation- Download all files specified in
files_to_download.txtto this directory - Create a directory called "index" and generate an index of the reference genome using the command
minimap2 -d index/hg38.mmi hg38.fa
- Run
run_subsample_pipeline.sh
We implement a baseline exact match filter using SIMD operations integrated in minimap2.
- For installation, run
make - General usage
minimap2 -d ref.mmi ref.fa # indexing
minimap2 -a ref.mmi reads.fq > alignment.sam # alignment
For more information about minimap2, please refer to its original repo.
- We implement the exact match filer in
exact2_match_sse.c - The filter in used in
map.cby calling functionexact_match_sse - If a read is detected to be an exact match, the mapper skips the expensive alignment step performed in
ksw_extz2_sse
Software GenStore is an implementation of the GenStore filter without in-storage support.
- Set the environment variables
REF_FILE,READ_FILE,HASH_SIZE,LOG2_NUM_THREADS. For example, to use the provided sample data, set the variables as follows:
REF_FILE=sample_data/NC_000913.3.head1000.fa
READ_FILE=sample_data/reads.fq
HASH_SIZE=48
LOG2_NUM_THREADS=2
- Compile the hash sorter and minimap 2 by running
makeingenstore-sw-filterandgenstore-sw-filter/minimap2/
- Generate logs for the reference using the command
minimap2/minimap2 -w1 -k150 -d $REF_FILE.mmi $REF_FILE >$REF_FILE.log 2>/dev/null
- Generate a hash and position table for the reference by running
./gen_hash $REF_FILE.log > $REF_FILE.hashes
- Reduce the table to the target hash size using
./generate_index $HASH_SIZE $REF_FILE.hashes > $REF_FILE.$HASH_SIZE.hashes.bin
- Index the table using
./index_index $HASH_SIZE $REF_FILE.$HASH_SIZE.hashes.bin $LOG2_NUM_THREADS > $REF_FILE.$HASH_SIZE.hashes.bin.index
- Generate logs for the read file using the command
minimap2/minimap2 -w1 -k$READ_LENGTH -d $READ_FILE.mmi $READ_FILE >$READ_FILE.log 2>/dev/null
- Generate a table for the reads by running
./generate_read_hashes.sh $READ_FILE.log > $READ_FILE.hashes
- Reduce the table to the target hash size using
./generate_reads $READ_LENGTH $HASH_SIZE $READ_FILE.hashes > $READ_FILE.$HASH_SIZE.hashes
- Index the table using
./index_reads $HASH_SIZE $READ_FILE.$HASH_SIZE.hashes $LOG2_NUM_THREADS > $READ_FILE.$HASH_SIZE.hashes.index
- Run the filter using
./check_files_mt $HASH_SIZE $REF_FILE.$HASH_SIZE.hashes.bin $READ_FILE.$HASH_SIZE.hashes
For example, for the provided input set, the output should look like the following:
bit width: 48 num_threads: 4
69782 1001 725 0.724276
where 0.724276 is the ratio of total reads that exactly match some subsequences in the reference genome.
We evaluate hardware configurations using two state-of-the-art simulators to analyze the performance of GenStore. We model DRAM timing with the DDR4 interface in Ramulator, a widely-used, cycle-accurate DRAM simulator. We model SSD performance using MQSim, a widely-used simulator for modern SSDs. We model the end-to-end throughput of GenStore based on the throughput of each GenStore pipeline stage: accessing NAND flash chips, accessing internal DRAM, accelerator computation, and transferring unfiltered data to the host.
We implement GenStore's accelerator units in Verilog to faithfully measure the throughput of the accelerators, and their area and power cost. We use Design Compiler version N-2017.09. The implementation can be found in genstore-hdl folder.
- In
key-script-command.tcl,path_to_verilog_filesis the path to genstore verilog source files,<verilog_module>.vis the file name containing the verilog module to synthesize, and<verilog_module_name>is the name of the module defined in this verilog file - Open up Synopsys command line
- Run
key-script-command.tcl
We will soon release the scripts used for Ramulator to model DRAM timing and the scripts used for MQSim to model SSD timing.
We will soon release the script used for modelling the end-to-end throughput of GenStore based on the throughput of each GenStore pipeline stage.
Nika Mansouri Ghiasi - n.mansorighiasi@gmail.com