This software is a non-official implementation of the paper "The Wasserstein Distance as a Dissimilarity Measure for Mass Spectra with Application to Spectral Deconvolution" by S. Majewski, M. A. Ciach, M. Startek, W. Niemyska, B. Miasojedow and A. Gambin.
Default solver is based on a problem-specific genetic algorithm. This allows for numerical stability and high speed.
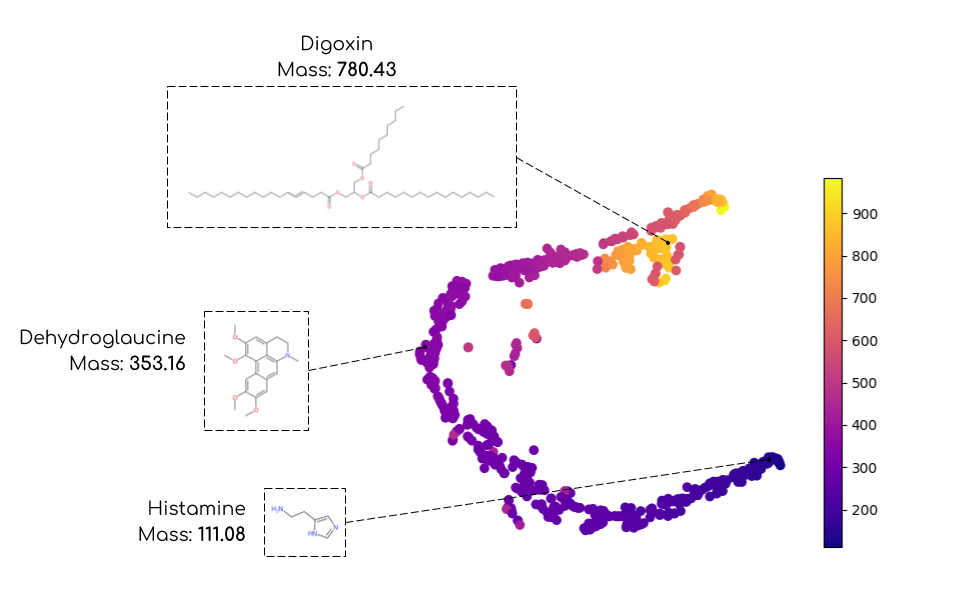 Figure: t-distributed Stochastic Neighbor Embedding of MS1 ESI-QTOF spectra
with precomputed matrix Wasserstein distances. The colormaps
gives the mass of each molecule (point) expressed in Daltons.
Figure: t-distributed Stochastic Neighbor Embedding of MS1 ESI-QTOF spectra
with precomputed matrix Wasserstein distances. The colormaps
gives the mass of each molecule (point) expressed in Daltons.
Run mass spectral deconvolution algorithm on an empirical spectrum and a folder containing the theoretical spectra. Paths to theoretical spectra should be specified in molecule_list.txt. A concrete example of dataset is provided in example-msd/ folder.
deconvms path/to/empirical/spectrum.txt path/to/molecule_list.txt path/to/molecules/folder --niter 50000
Compute Wasserstein dissimilarity between two mass spectra:
wassms path/to/first/record/file.txt path/to/second/record/file.txt --m W
Compute Euclidean distance between two mass spectra:
wassms path/to/first/record/file.txt path/to/second/record/file.txt --m E
Compile the project with g++:
make release
This will create the two executable files wassms and deconvms.
The project is built on top of Eigen, whose path is assumed to be stored in a Eigen3_DIR variable.