lncGSEA: an R package to infer lncRNA associated pathways from large-scale cancer transcriptome sequencing data
lncGSEA is a convenient tool to predict the lncRNA associated pathways through Gene Set Enrichment Analysis (GSEA) of gene expression profiles from large-scale cancer patient samples.lncGSEA is developed at Rendong Yang Lab at The Hormel Institute, University of Minnesota.
Workflow
Prerequisites
Install required packages
install.packages("devtools")
install.packages("data.table")
install.packages(c("dplyr", "stringr","tidyr","tibble","ggplot2"))
install.packages(c("ggrepel", "RColorBrewer"))
devtools::install_github("ctlab/fgsea")
Load required packages
library(devtools)
library(fgsea)
library(data.table)
library(tibble)
library(dplyr)
library(stringr)
library(tidyr)
library(ggplot2)
library(ggrepel)
library(RColorBrewer)
Install lncGSEA
install_github("ylab-hi/lncGSEA")
library(lncGSEA)
Download required datasets
There are two kinds of datasets required for lncGSEA to perform its function of finding enriched pathways regulated by lncRNAs.
- lncRNA expression in human cancer samples.
- Two public databases: mitranscriptome beta and RefLnc. The files were named as "mitranscriptome.expr.fpkm.tsv.gz" and "RefLnc_lncRNA_tumor_sample_FPKM.gz", respectively.
- gene expression matrix (FPKM) for each cohort in TCGA study.
- Example: PRAD.FPKM.txt, BRCA.FPKM.txt, COAD.FPKM.txt
All datasets can be downloaded from this shared link: https://drive.google.com/drive/folders/1g3Rl4I5RA2Xf_7u6Y3IBKWCIDYf-w99b?usp=sharing
Create a data folder
Please create a data folder in your current working directory by the following command to store the downloaded datasets.
if (!file.exists("data")){
dir.create("data")
}
Examples
1. Make the dataset ready for lnc_gsea
Create an expression data frame for PRAD cohort. The first column of output should be the cohort name, the first argument of pre_gsea and the second column name should be one of the transcript ids of the interested lncRNA (e.g., ENST00000561519 of ARLNC1), other columns are genes from the specific cohort which passed the expression threshold (e.g., mean of FPKM larger than 1). Rows are tumor samples from PRAD cohort. A same transcript id may have different versions, pre_gsea use transcript id without version number ("ENST00000561519" instead of "ENST00000561519.[0-9]").
test <- pre_gsea("PRAD", "ENST00000561519", "./data/")
test[1:5, 1:5]
The first 5 rows and columns are shown below:
PRAD ENST00000561519.5 TSPAN6 DPM1 SCYL3
1754 TCGA-G9-7521-01A-11R-2263-07 0.0104 10.62 24.92 1.971
1757 TCGA-KK-A7AU-01A-11R-A32O-07 0.2343 13.35 23.39 3.539
1760 TCGA-EJ-7125-01A-11R-1965-07 0.7116 18.71 15.37 1.835
1763 TCGA-ZG-A9LM-01A-11R-A41O-07 0.4569 16.89 29.95 2.484
1765 TCGA-QU-A6IM-01A-11R-A31N-07 0.0000 8.45 9.04 0.789
2. Run lnc_gsea function on output test
The pathway enrichement analysis is implemented by fgsea function from R package "fgsea". (https://github.com/ctlab/fgsea)
The default ranking metric is "pearson" correlation coefficient, you can also set cor.method = "spearman" to apply "spearman" correlation coefficient as ranking metric. The other ranking metric is "logFC", which is log2FoldChange between high expressed lncRNA group vs low expressed group.
The default geneset is NULL, in this case, lnc_gsea will use HALLMARK gene set from MSigDB. You can provide your own customized gene set too, for example, your gene set is stored in a folder called "gmt" at current working directory, geneset can be set as "./gmt/yourgeneset.gmt".
You can also set genelist = TRUE, to save a ranked gene list data frame for pre-ranked GSEA analysis using GSEA desktop app from Broad Institute. This ranked gene list data frame has two columns, the first column is gene name, the second column is ranking metric, either logFC or correlation coefficient in decreasing order. The main output of this function is the enriched pathways ranked by NES (normalized enrichement score) in a descending order. If you want to visualize an interested pathway, set pathway = "you pathway", an enrichement plot can be saved in your current working directory.
lnc_gsea(tid_cohort = test, metric = "cor", cor.method = "pearson", genelist = TRUE, geneset = NULL, pathway = NULL)
The first 4 rows output of lnc_gsea is shown as below:
pathway pval padj ES NES nMoreExtreme size leadingEdge
HALLMARK_ANDROGEN_RESPONSE 0.004950 0.011421 0.557500 2.645172 0 94 ABCC4|RPS6KA3|SMS|PDLIM5|ELL2|ALDH1A3
HALLMARK_FATTY_ACID_METABOLISM 0.005319 0.011563 0.414583 2.0858619 0 135 ACADL|SMS|BPHL|AADAT|MCEE|ACADM
HALLMARK_OXIDATIVE_PHOSPHORYLATION 0.007299 0.013924 0.365339 1.898366 0 182 GLUD1|ACADM|ACADSB|ALDH6A1|MPC1|GOT2
HALLMARK_UNFOLDED_PROTEIN_RESPONSE 0.005025 0.011421 0.394156 1.897353 0 106 SLC1A4|TUBB2A|WIPI1|PREB|PDIA5|SSR1
....
The first few rows ranked gene list data frame looks like below:
gene_name ranks
RP11.314O13.1 0.672201572333335
SLC4A4 0.463208163284544
RP11.114M1.1 0.457428137420679
NUDT4 0.428204695416086
LINC00578 0.417863932304566
MPC2 0.405784680718824
TRPM8 0.403574189464877
ABCC4 0.401451566178376
AL589822.1 0.392845646974538
TRIM36 0.38249804158695
CLGN 0.380270202148028
POTEH.AS1 0.379078513980204
MEAF6 0.368165483665729
GLYATL1 0.363674770886263
CITED2 0.36312799821486
....
3. Visualization
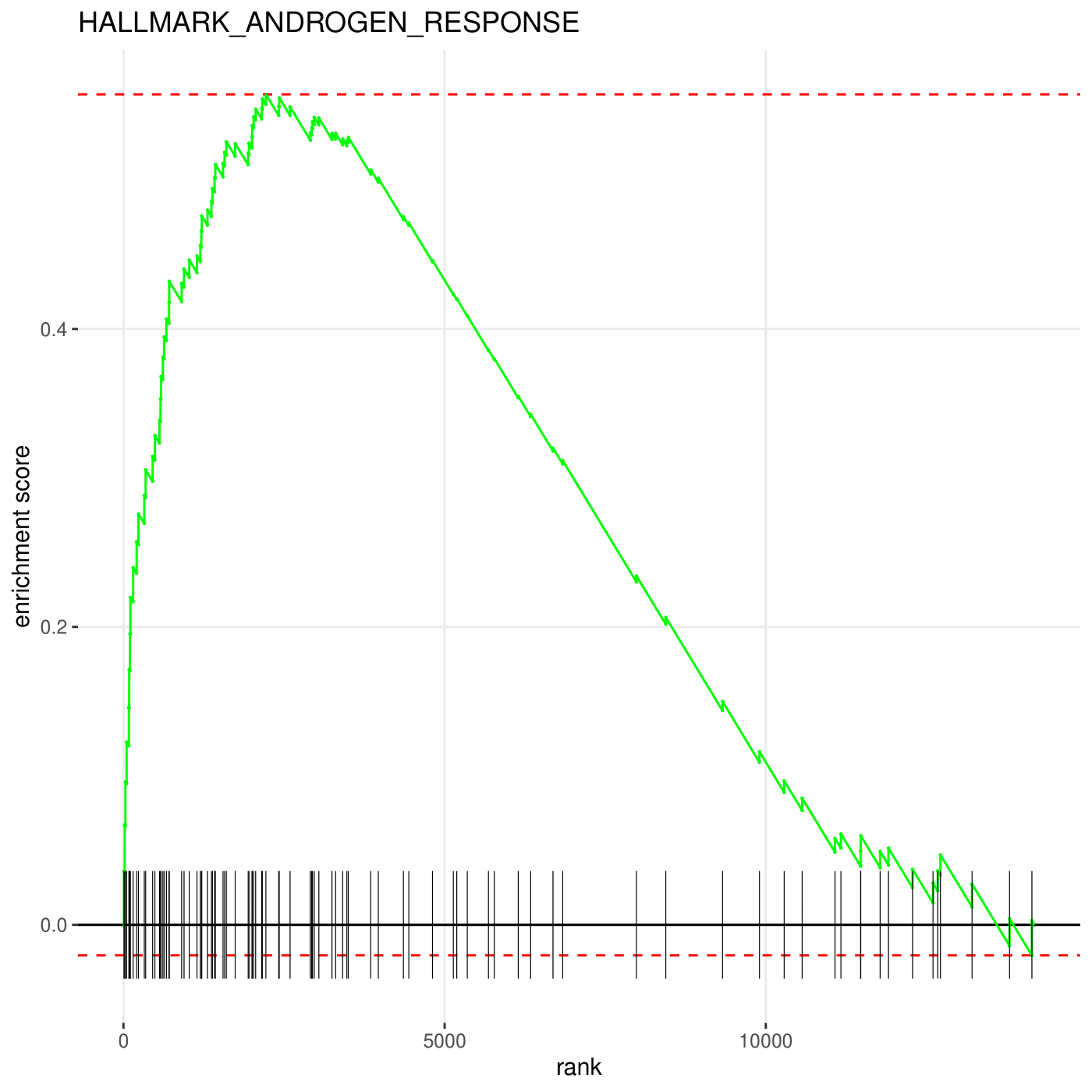
By setting pathway = "HALLMARK_ANDROGEN_RESPONSE" in lnc_gsea, you can have an enrichement plot of AR pathway, which is displayed as below. The plot is produced by plotEnrichment function of fgsea package.
3.1 Visualize enriched pathway results by applying plot_gsea to a saved *.txt output from lnc_gsea
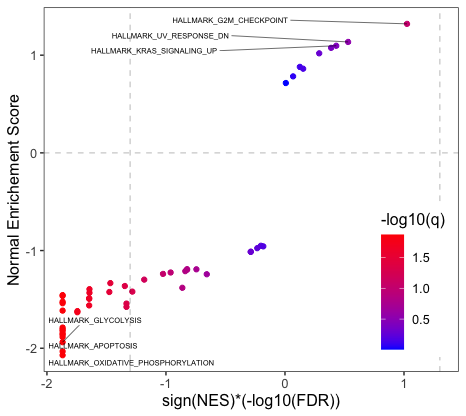
One can provide a customized gene pathways to be labelled in the plot by setting pathway.list = "pathway you want to label", the name of the gene pathways should be the same as the pathways in the gene set gmt file you have used in lnc_gsea. For example, if you want to label "HALLMARK_ANDROGEN_RESPONSE", you can set pathway.list = "HALLMARK_ANDROGEN_RESPONSE", or if you want to label multiple pathways, you can set pathway.list = c("HALLMARK_ANDROGEN_RESPONSE", "HALLMARK_FATTY_ACID_METABOLISM"). If pathway.list = NULL, by default, it will label top/bottom 3 pathways. You can choose how many pathways you want to label by n, by default n = 3. You also have the flexibility of choosing positive ("pos") or negative ("neg") or both ("both") enriched pathways to label.
plot_gsea("ENST00000561519.5_PRAD_cor.txt", direction = "both")
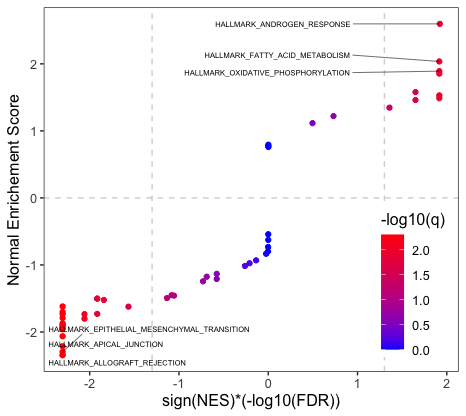
3.2 Compare one lncRNA's regulated pathways in different studies
If you have not run lnc_gsea for your interested lncRNA in multiple studies, you can obtain those results by running pre_compareCohort function, the output of this function can be directly used by function plot_compareCohort, which will produce a plot shown below. If you already have enriched pathways results for the interested lncRNA in multiple studies, you can apply function pre_multiCompare and set compare = "cohort", and then feed the output to plot_compareCohort to obtain plot like below.
Suppose one wants to compare the difference of enriched pathways for transcript "ENST00000561519" of ARLNC1 in prostate, lung and breast cancer. Here are the two ways to generate the data frame for the comparison plot.
# run from the scratch
cohorts <- c("PRAD","LUAD","BRCA")
arlnc1.df <- pre_compareCohort(lncRNA="ENST00000561519", cohorts = cohorts, pathtofile="./data/")
# already have lnc_gsea output
arlnc1.df <- pre_multiCompare(files = list("ENST00000561519_PRAD_cor.txt",
"ENST00000561519_LUAD_cor.txt",
"ENST00000561519_BRCA_cor.txt"),
compare = "cohort")
plot_multiCompare(arlnc1.df)
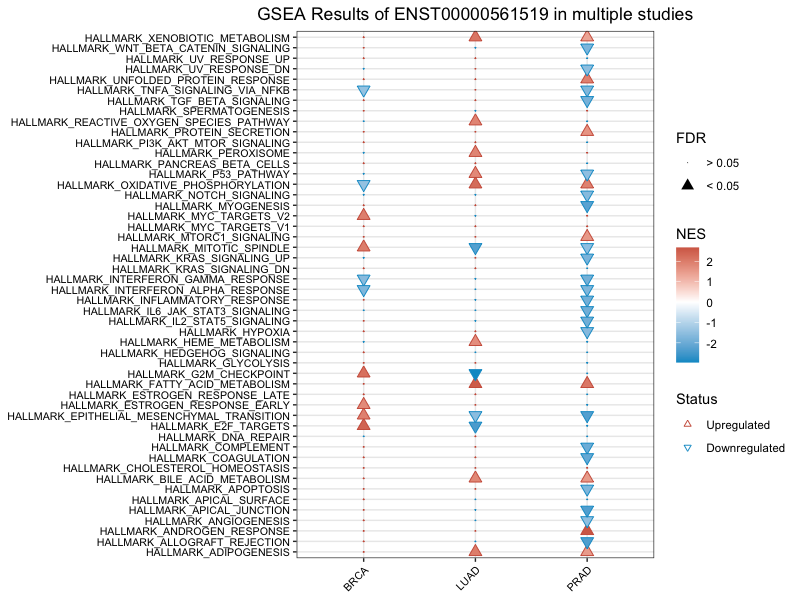
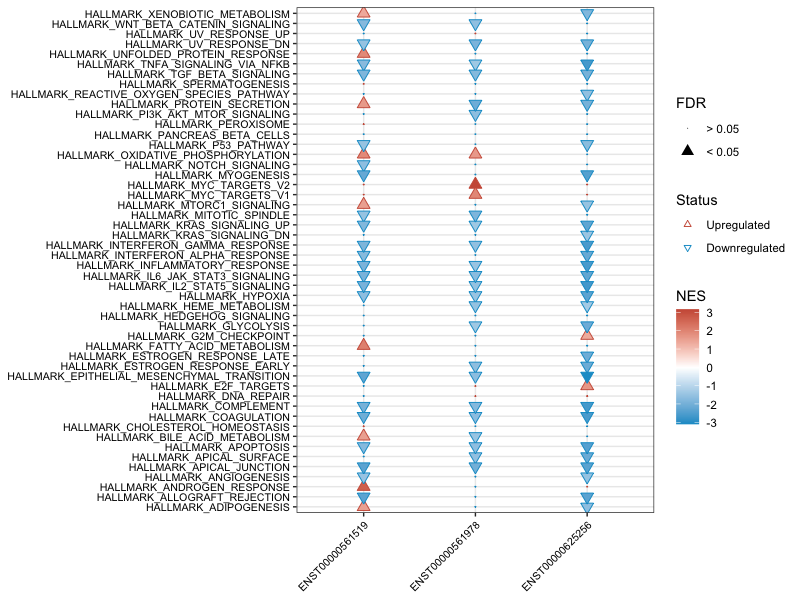
3.3 Study multiple lncRNAs' regulatory pathway in one cancer
Similarly, you can collectively compare multiple lncRNAs regulatory enriched pathway in one cancer. Here suppose you already have results from lnc_gsea for the list of interested lncRNAs. You can obtain a similar plot by running the following functions:
prad <- pre_multiCompare(files = list("ENST00000625256_PRAD_cor.txt",
"ENST00000561978_PRAD_cor.txt",
"ENST00000561519_PRAD_cor.txt"),
compare = "lncRNA")
plot_multiCompare(prad)
An example of lncRNA expression matrix provided by user
If you have an interested lncRNA which has not been included in those two databases yet, you can provide your own customized lncRNA expression matrix in TCGA cohorts.
An example of the customized lncRNA expression data frame or matrix should look like:
TCGA-ZG-A9LM-01A-11R-A41O-07 TCGA-V1-A8WW-01A-11R-A37L-07 ...
PB.69 0.0632 0.0234 ...
Example of other genes' expression data should look like below:
TCGA-ZG-A9LM-01A-11R-A41O-07 TCGA-V1-A8WW-01A-11R-A37L-07 ...
TSPAN6 16.892 12.081 ...
DPM1 29.951 25.948 ...
SCYL3 2.484 4.688 ...
FGR 0.956 0.899 ...
CFH 2.961 1.490 ...
The most important tip for combining these two data frames by custom_lnc is the column names from both data frames should be the same for the same person. However, the order of the columns or the numbers of the columns can be different.
lnctest <- custom_lnc("./data/lncRNA.custom.txt","./data/PRAD.FPKM.txt")
lnc_gsea(tid_cohort = lnctest, metric = "cor",cor.method = "pearson")
plot_gsea("PB.69_PRAD_cor.txt")
License
The project is licensed under the MIT license.
Contact
Bug reports or feature requests can be submitted on the lncGSEA Github page.
Citation
Ren Y., Wang TY., Anderton LC., Cao Q. and Yang R. lncGSEA: an R package to infer lncRNA associated pathways from large-scale cancer transcriptome sequencing data. (Submitted)