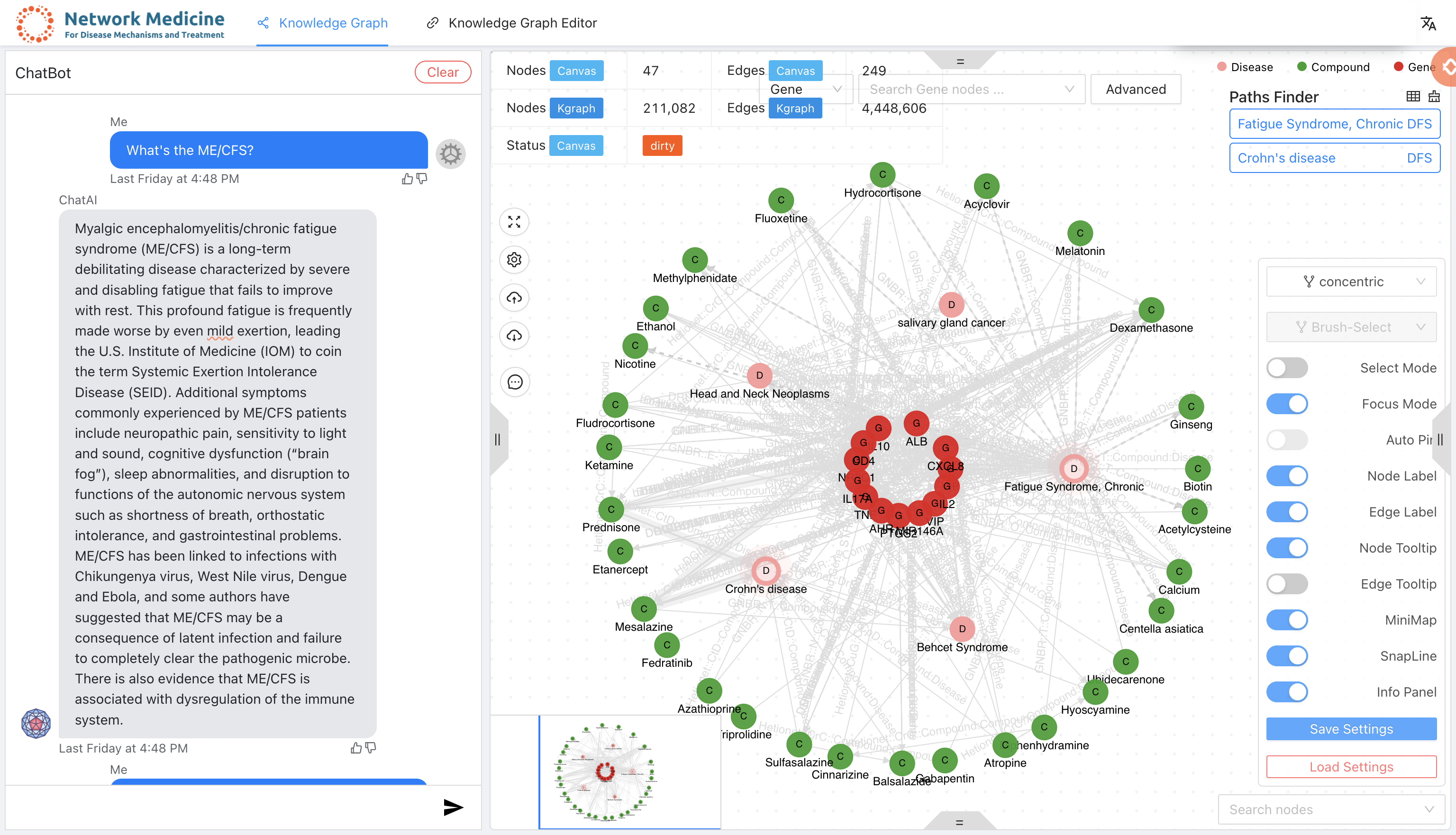
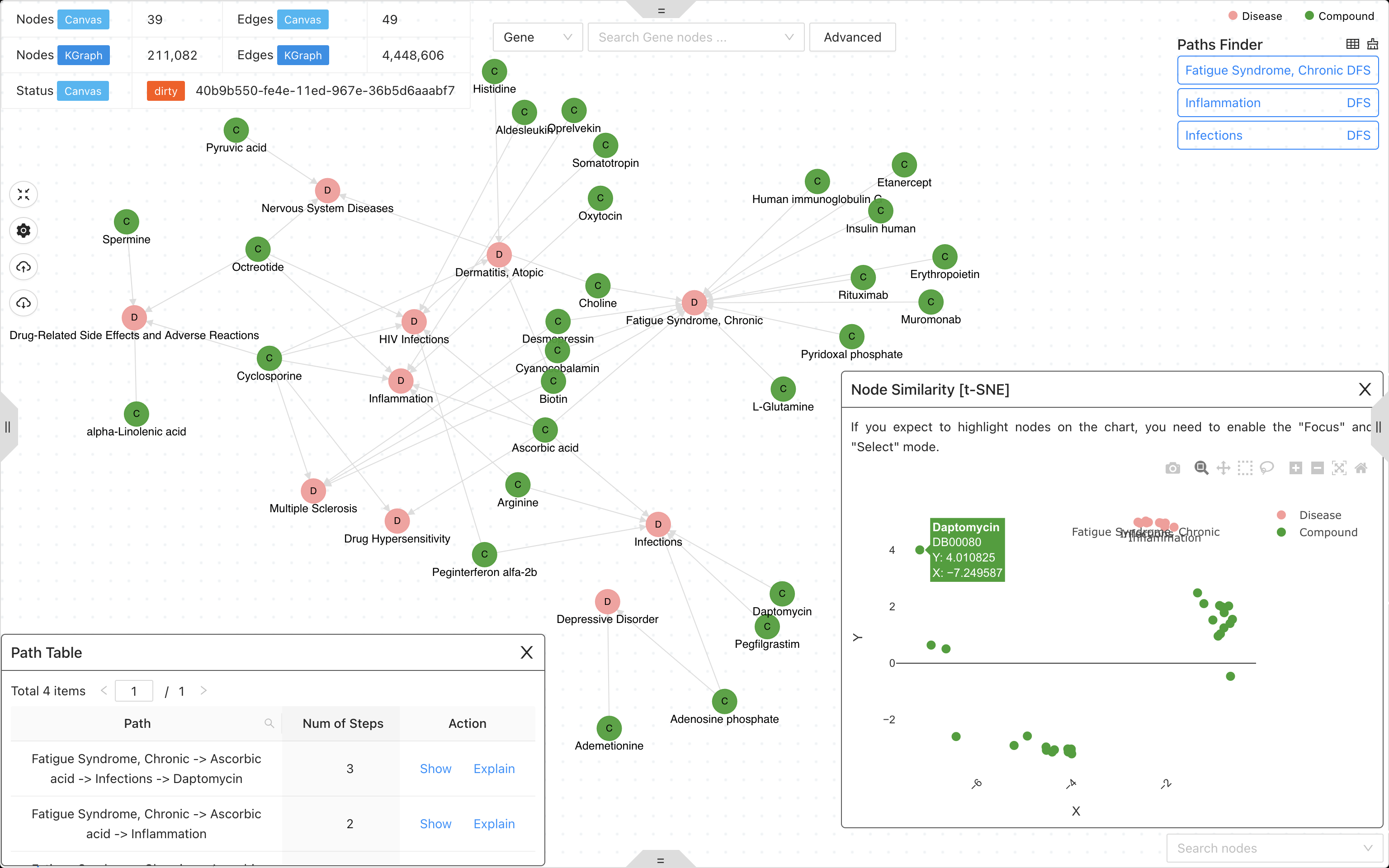
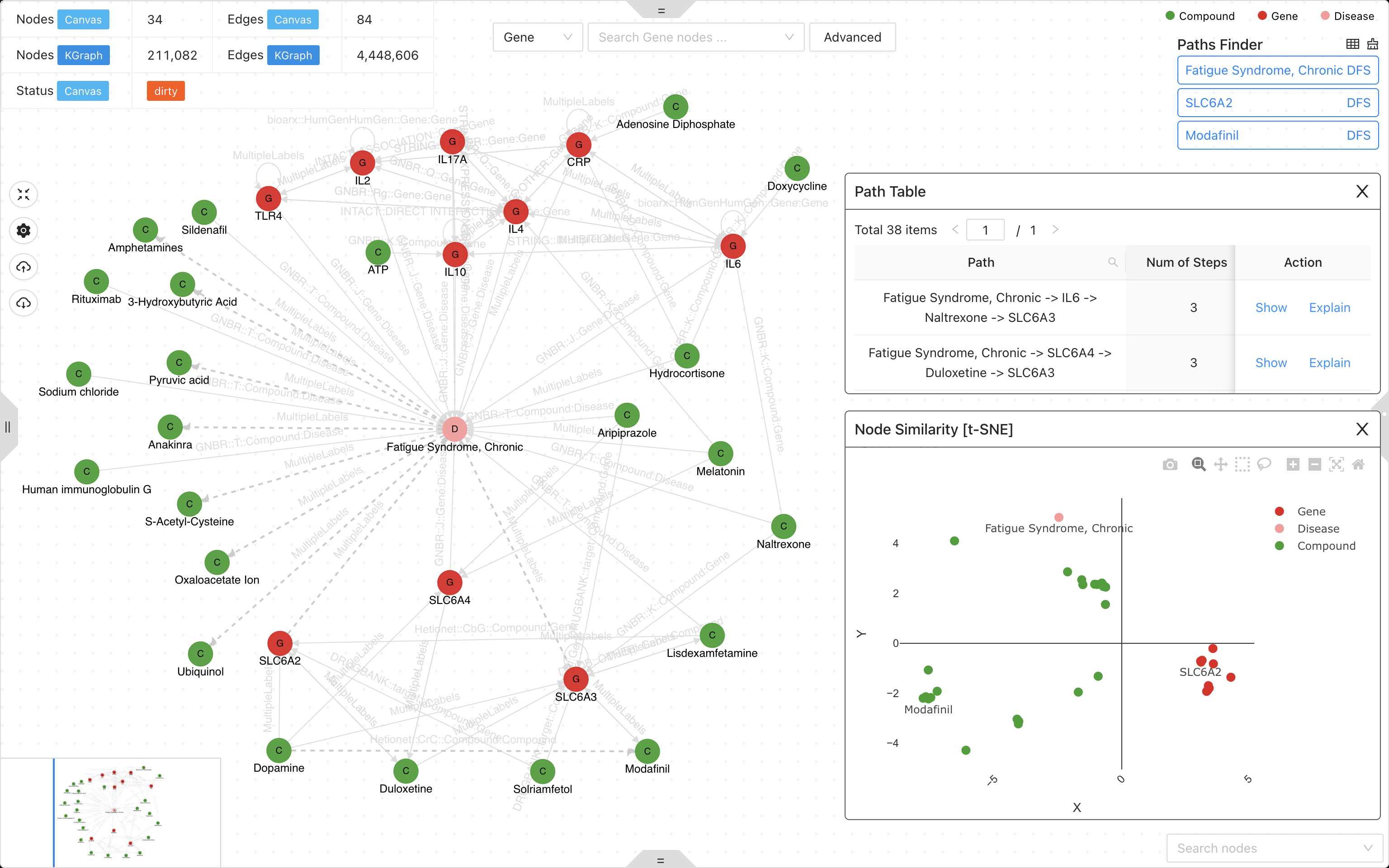
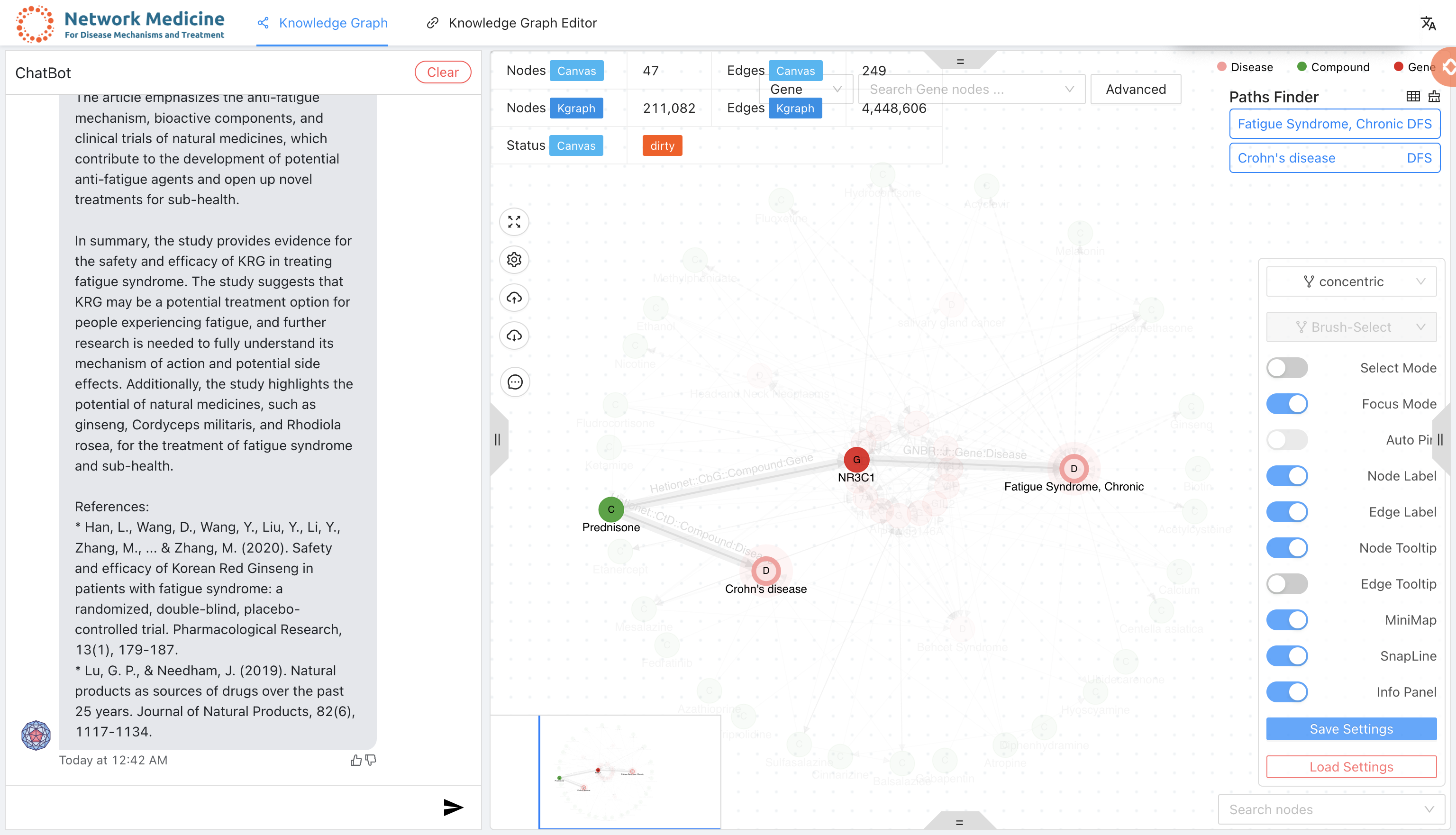
A knowledge graph system with graph neural network for drug discovery, disease mechanism, biomarker screening and discovering response to toxicant exposure.
- Knowledge graph studio for graph query, visualization and analysis.
- Graph neural network for drug discovery, disease mechanism, biomarker screening and discovering response to toxicant exposure.
- Support customized knowledge graph schema and data source.
- Support customized graph neural network model.
- Support customized omics datasets.
- Integrated large language models (such as vicuna, rwkv, chatgpt etc. more details on chat-publications) for answering questions.
git clone https://github.com/yjcyxky/rapex.git./bin/lein uberjarbash create-db.sh rapex_dev 54320
export MIGRATE_DB=true
export JAVA_OPTS='-Dconf=./example-conf.edn'
bash bin/startcd studio && yarn start:local-dev
# Release it to GitHub
cd studio && yarn build && yarn gh-pagesFIXME: explanation
$ java -jar rapex-0.1.0-standalone.jar [args]
FIXME: listing of options this app accepts.
...
- [] Add advanced search for knowledge graph.
- [] Two modes: add to current canvas & add to new canvas.
Copyright © 2022 FIXME
This program and the accompanying materials are made available under the terms of the Eclipse Public License 2.0 which is available at http://www.eclipse.org/legal/epl-2.0.
This Source Code may also be made available under the following Secondary Licenses when the conditions for such availability set forth in the Eclipse Public License, v. 2.0 are satisfied: GNU General Public License as published by the Free Software Foundation, either version 2 of the License, or (at your option) any later version, with the GNU Classpath Exception which is available at https://www.gnu.org/software/classpath/license.html.