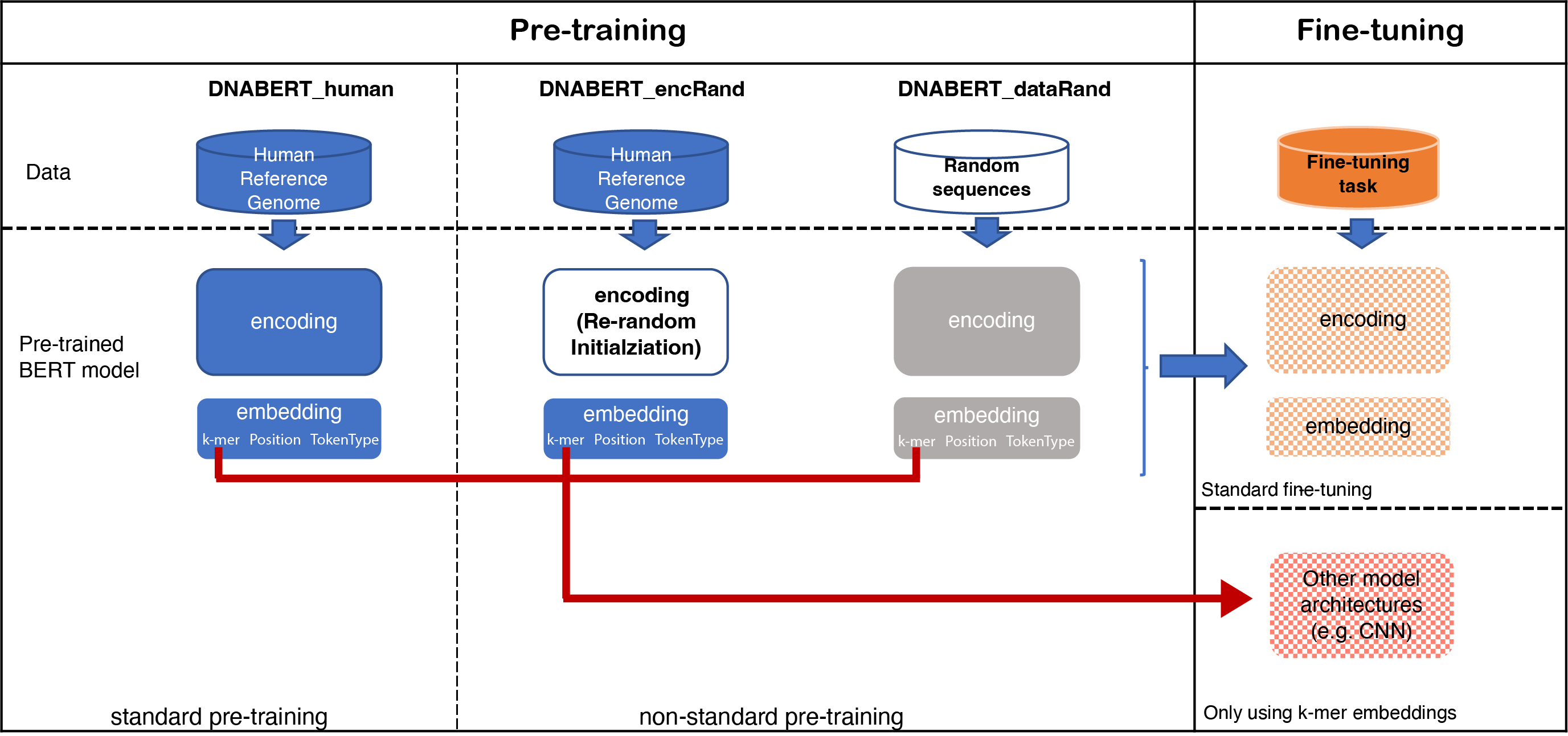
Investigation of the BERT model on nucleotide sequences with non-standard pre-training and evaluation of different k-mer embeddings
In this study, we used a non-standard pre-training approach through incorporating randomness at the data and model level to investigate a BERT model pre-trained on nucleotide sequences.
- data/ptData: source code of generate random sequences.
- TATA: human and mouse TATA dataset are in the fold of data/ftData/TATA
- TFBS: used motif_discovery(690) and motif_occupancy (422) curated dataset provided by Zeng et al. (https://academic.oup.com/bioinformatics/article/32/12/i121/2240609). Please use the paper provided URL for the download.
- ft_tasks: source code of using different k-mer embeddings in downstream tasks of TATA prediciton and TBFS prediction.
| k-mer embedding | Description | required files |
|---|---|---|
| dnabert | k-mer embedding from DNABERT pre-trained on hg38 | pre-trained model provided by DNABERT |
| dnabert | k-mer embedding from DNABERT pre-trained on random data | DNABERT model pre-trained on random data |
| onehot | one-hot embedding | None |
| dna2vec | k-mer embedding from dna2vec | pretrained model |
KMER=5
SPIECE= "human" (or "mouse")
MODEL="deepPromoterNet"
MODEL_SAVE_PATH="model/"
DATA_PATH="ftData/TATA/TATA_${SPIECE}/overall"
EMBEDDING="dnabert" (or "onehot", "dna2vec")
embed_file=FOLD_PATH_OF_THE_PRETRAINED_MODEL (or NONE)
KERNEL="5,5,5"
LR=1e-4
EPOCH=20
BS=64
DROPOUT=0.1
CODE="ft_tasks/TATA/tata_train.py"
python $CODE --kmer $KMER --cnn_kernel_size $KERNEL --model $MODEL --model_dir $MODEL_SAVE_PATH \
--data_dir $DATA_PATH --embedding $EMBEDDING --embedding_file $embed_file \
--lr $LR --epoch $EPOCH --batch_size $BS --dropout $DROPOUT --device "cuda:0"
KMER=5
MODEL="zeng_CNN"
KERNEL="24"
MODEL_SAVE_PATH="model/"
DATA_PATH="TBFS/motif_discovery/" or "TBFS/motif_occupancy/"
EMBEDDING="dnabert" (or "onehot", "dna2vec")
embed_file=FOLD_PATH_OF_THE_PRETRAINED_MODEL (or NONE)
LR=0.001
EPOCH=10
BS=64
DROPOUT=0.1
CODE="ft_tasks/TFBS/TBFS_all_run.py"
python $CODE --kmer $KMER --cnn_kernel_size $KERNEL --model $MODEL --model_dir $MODEL_SAVE_PATH \
--data_dir $DATA_PATH --embedding $EMBEDDING --embedding_file $embed_file \
--lr $LR --epoch $EPOCH --batch_size $BS --dropout $DROPOUT --device "cuda:0"
-
pt_models: download link of the pre-trained model using random data.
-
4mer pre-trained on randomly generated sequences: https://drive.google.com/file/d/1YKKoX_8NRrPR13uGdEQAKWqxBcOvq2su/view?usp=share_link
-
5mer pre-trained on randomly generated sequences: https://drive.google.com/file/d/1a2OjubusbsXkC2xAp8W0BbVZHuqyCAhk/view?usp=share_link
-
6mer pre-trained on radomly generated sequences: https://drive.google.com/file/d/1-6XMO70jY9Tdj9R19vgq8u9DtCzDGPkm/view?usp=share_link
- results: detailed results of each dataset of TBFS tasks.