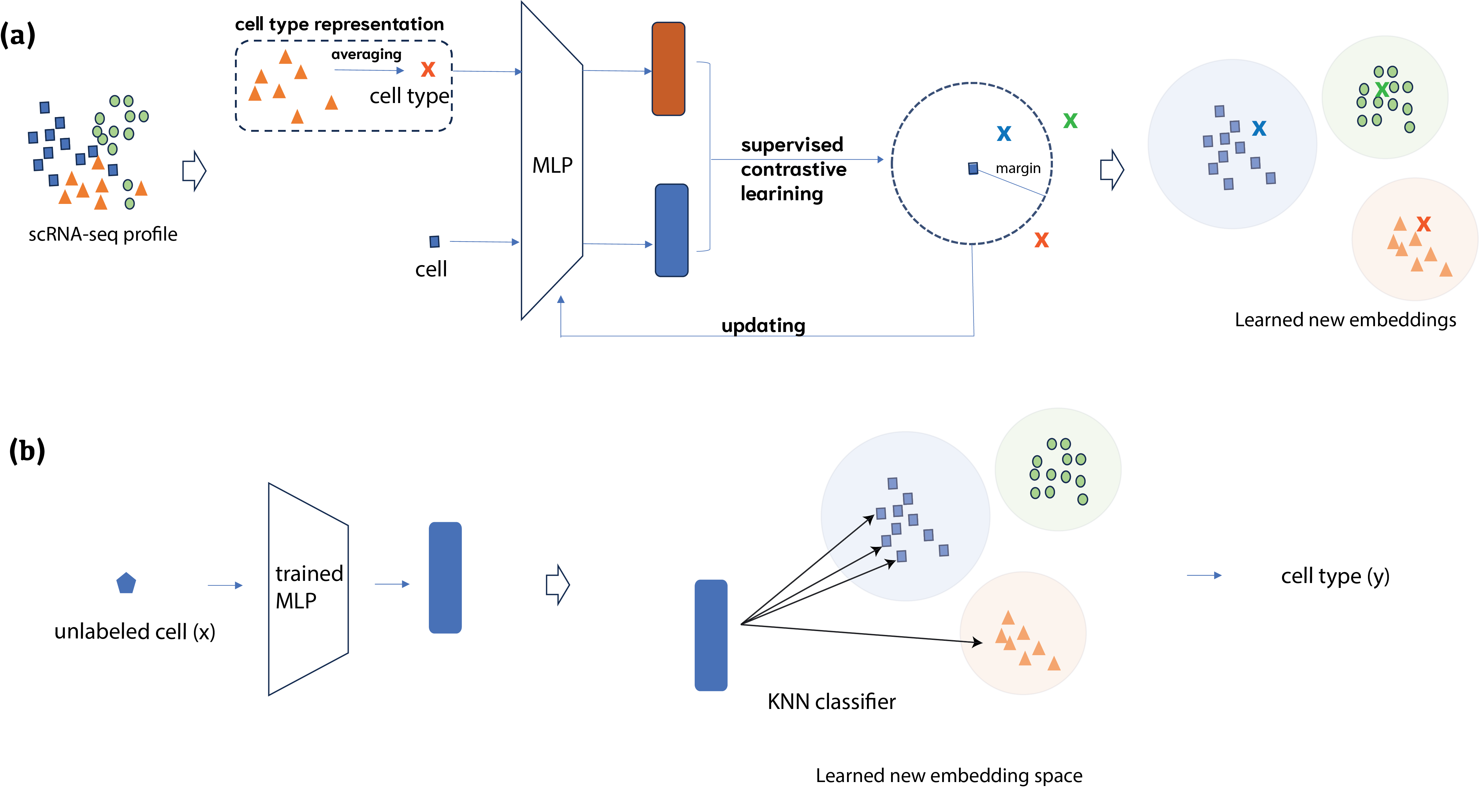
In this work, we developed a novel modeling formalism for cell type annotation with a supervised contrastive learning method, named SCLSC (Supervised Contrastive Learning for Single Cell). Different from the previous usage of contrastive learning in single cell data analysis, we employed the contrastive learning for instance-type pairs instead of instance-instance pairs. Mores specifically, in the cell type annotation task, the contrastive learning is applied to learn cell and cell type representation that render cells of the same type to be clustered in the new embedding space. Through this approach, the knowledge derived from annotated cells is transferred to the feature representation for scRNA-seq data.
- python 3
- Pytorch 1.11
- scanpy
- anndata
The datasets underwent preprocessing to eliminate cells with high mitochondrial gene expression (more than 5 percents of the cell total count), cells with minimal gene expression (number of genes per cell < 200), and genes that were only detected in a small number of cells (number of cells that expressed the gene < 3).
Subsequently, We selected 2000 highly variable genes (HGV) using analytic Pearson residuals implemented in Scanpy package.
Following this, we normalized the count of each cell to 10,000 counts and applied a
- PBMC: Fresh 68k PBMCs Donor A
- Pancreas: Figshare Pancreas Link
- Thymus: Thymus link
- Lung: Figshare Lung Link
- CeNGEN: CeNGEN Data on GitHub
- Zebrafish: Zebrafish Dataset
- Dengue dataset: EBI Dengue Dataset
ENCODER="MLP"
MARGIN=1.0
BATCH_SIZE=256
EPOCH=100
GENE_SET="hvg"
DATADIR=PATH_of_DATA_DIRECTORY
MODEL_SAVE_DIR=PATH_of_MODEL_SAVE_DIRECTORY
DATASET="zebrafish_all"
OUTPUT_DIR=PATH_of_OUTPUT_DIRECTORY
echo "Processing $DATASET"
python sc2l_main.py --encoder $ENCODER --dataset_name $DATASET --data_dir $DATADIR --model_dir $MODEL_SAVE_DIR \
--gene_set ${GENE_SET} --margin $MARGIN --output_dir $OUTPUT_DIR --epoch $EPOCH --batch_size $BATCH_SIZE
ENCODER="MLP"
MARGIN=1.0
BATCH_SIZE=256
EPOCH=100
GENE_SET="hvg"
DATASET="pancreas_all"
DATADIR=PATH_of_DATA_DIRECTORY
MODEL_DIR=PATH_of_SAVED_MODEL_DIRECTORY
KNN_K=10
echo "Processing $DATASET"
python sc2l_test_knn.py --encoder $ENCODER --dataset_name $DATASET --data_dir $DATADIR --model_dir $MODEL_DIR \
--gene_set ${GENE_SET} --margin $MARGIN --knn_k $KNN_K --epoch $EPOCH