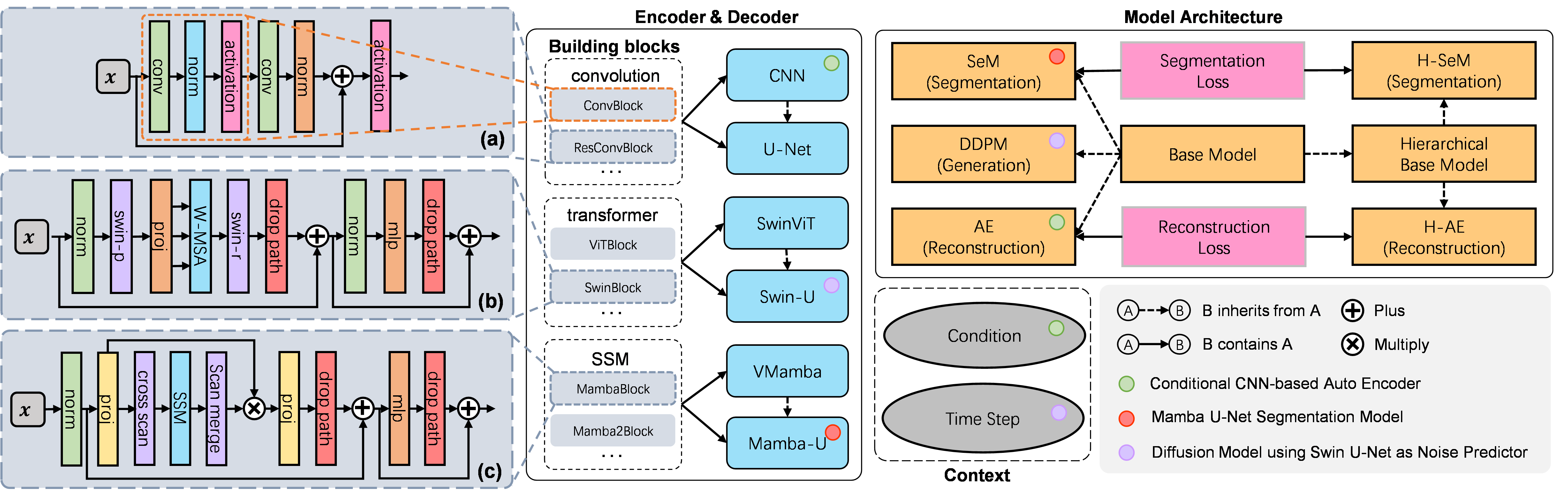
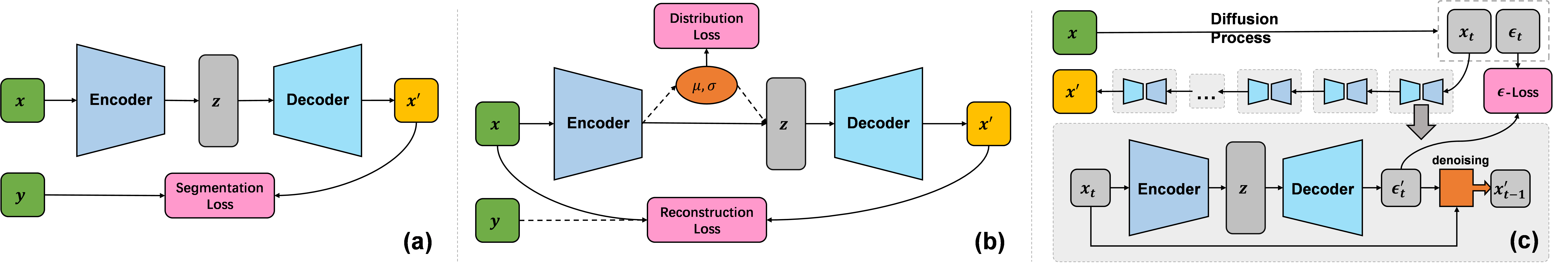
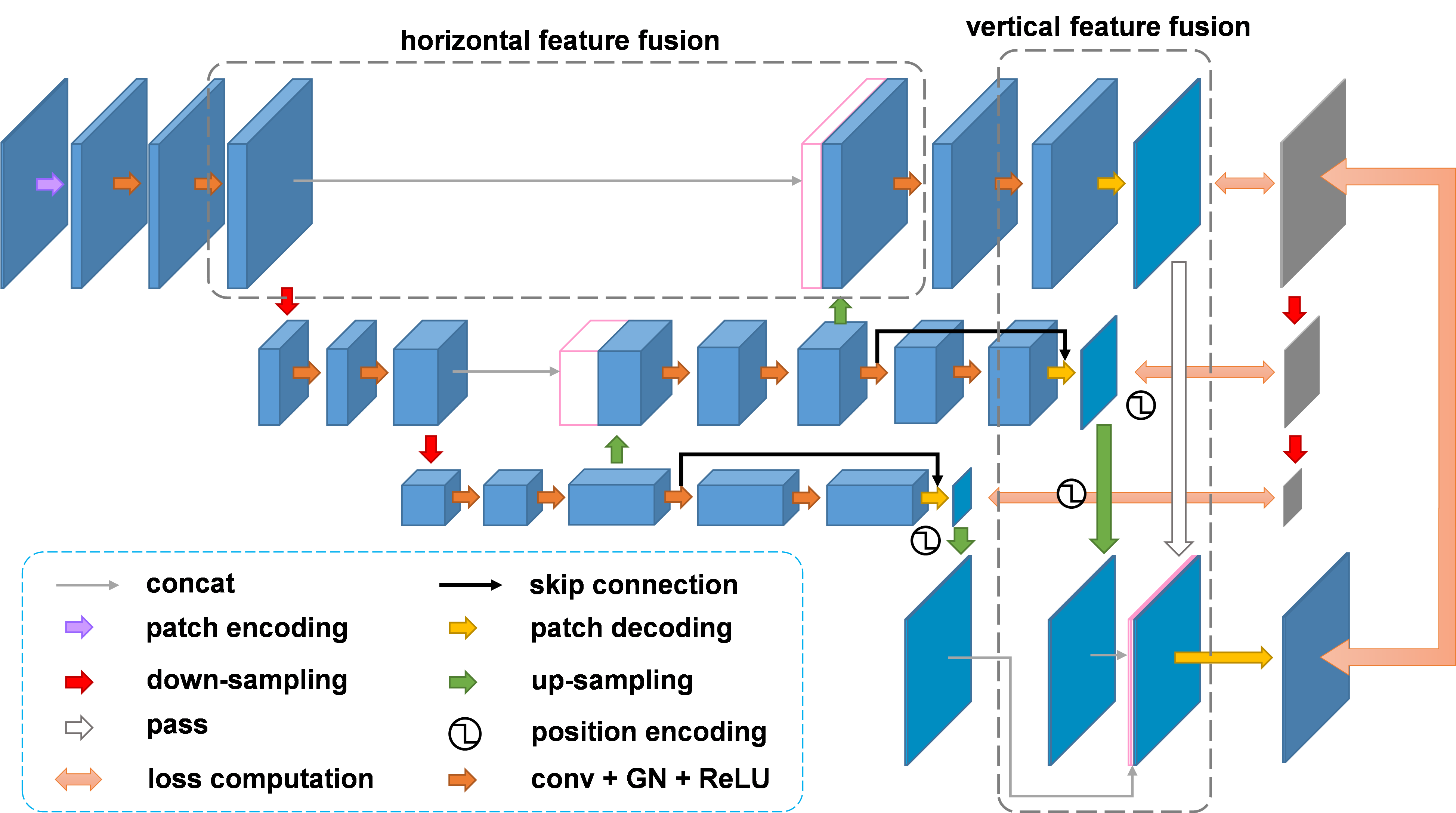
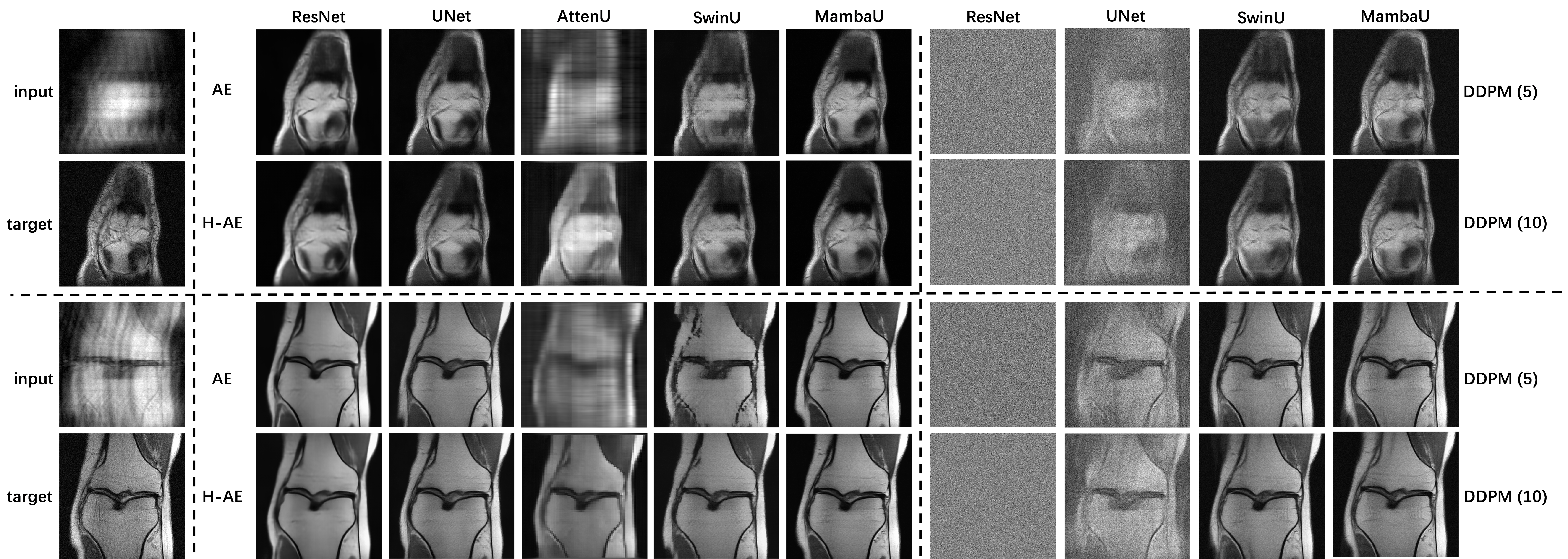
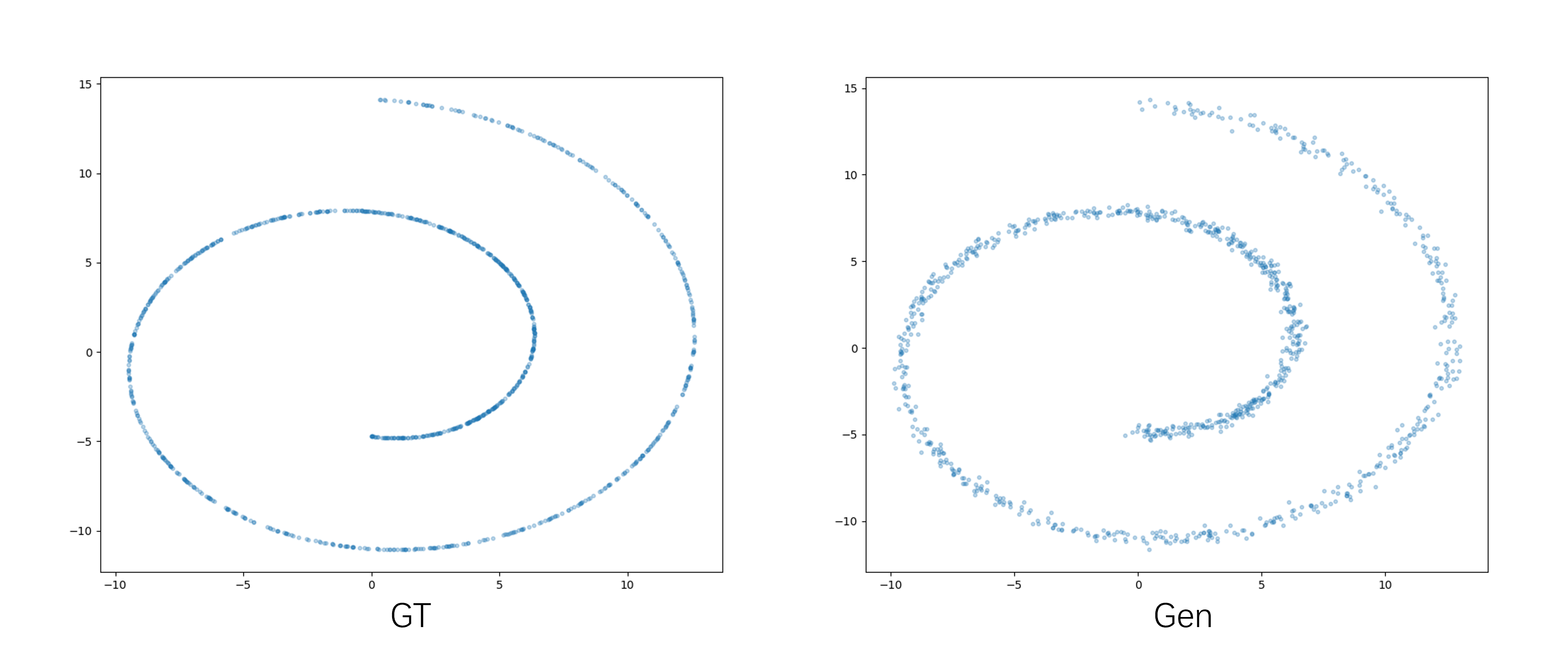
Flemme is a flexible and modular learning platform for medical images. In Flemme, we separate encoders from the model architectures, enabling fast model construction via different combinations for medical image segmentation, reconstruction and generation. In addition, a general hierarchical architecture with a pyramid loss is proposed for vertical feature refinement and integration.
We are also working on Flemme to support point cloud modeling.
torch torchvision simpleitk nibabel matplotlib scikit-image scikit-learn tensorboard
einops
mamba-ssm (CUDA version >= 11.6)
POT plyfile
You can modify flemme/config.py to disable some components of Flemme so that you don't need to install the corresponding required packages.
Git clone from git@github.com:wlsdzyzl/flemme.git
Run following commands in terminal to setup Flemme to your environment:
cd flemme
python setup.py install
Creating a deep learning model with Flemme is quite straightforward; you don't need to write any code. All things can be down through a yaml config file. An example of constructing a segmentation model with UNet encoder using ResConvBlock looks like:
model:
### architecture
name: SeM
### encoder
encoder:
name: UNet
image_size: [320, 256]
in_channel: 3
out_channel: 1
patch_channel: 32
patch_size: 2
down_channels: [64, 128, 256]
middle_channels: [512, 512]
building_block: res_conv
normalization: batch
### loss function
segmentation_losses:
- name: Dice
- name: BCEL
You may also need to specify the data-loader, optimizer, checkpoint path and other hyper-parameters. A full configuration refers to resources/img/biomed_2d/cvccdb/train_unet_sem.yaml. To train the model, run command:
train_flemme --config path/to/train_config.yaml
For visualization of the training process:
tensorboard --logdir path/to/ckp/
For testing:
test_flemme --config path/to/test_config.yaml
Supported encoders:
- [CNN, UNet, ViT, ViTU, Swin, SwinU, VMamba, VMambaU] for 2D/3D image
- [PointWise, PointNet, DGCNN] for point cloud.
A encoder named as XXU indicates it's a U-shaped encoder.
UNet is an alias of CNNU.
Supported Architectures:
- [SeM, HSeM] for segmentation,
- [AE, HAE, SDM] for reconstruction,
- [VAE, DDPM, DDIM, LDM] for generation.
DDIM refers to denoising diffusion implicit model, which is a fast sample strategy.
SDM refers to supervised diffusion model (use input as a input condition of ddpm).
LDM refers to latent diffusion model, constructed with a auto-encoder/ vae and ddpm.
A detailed instruction of supported encoders, context embeddings, model architectures and training process can refer to documentation of flemme.
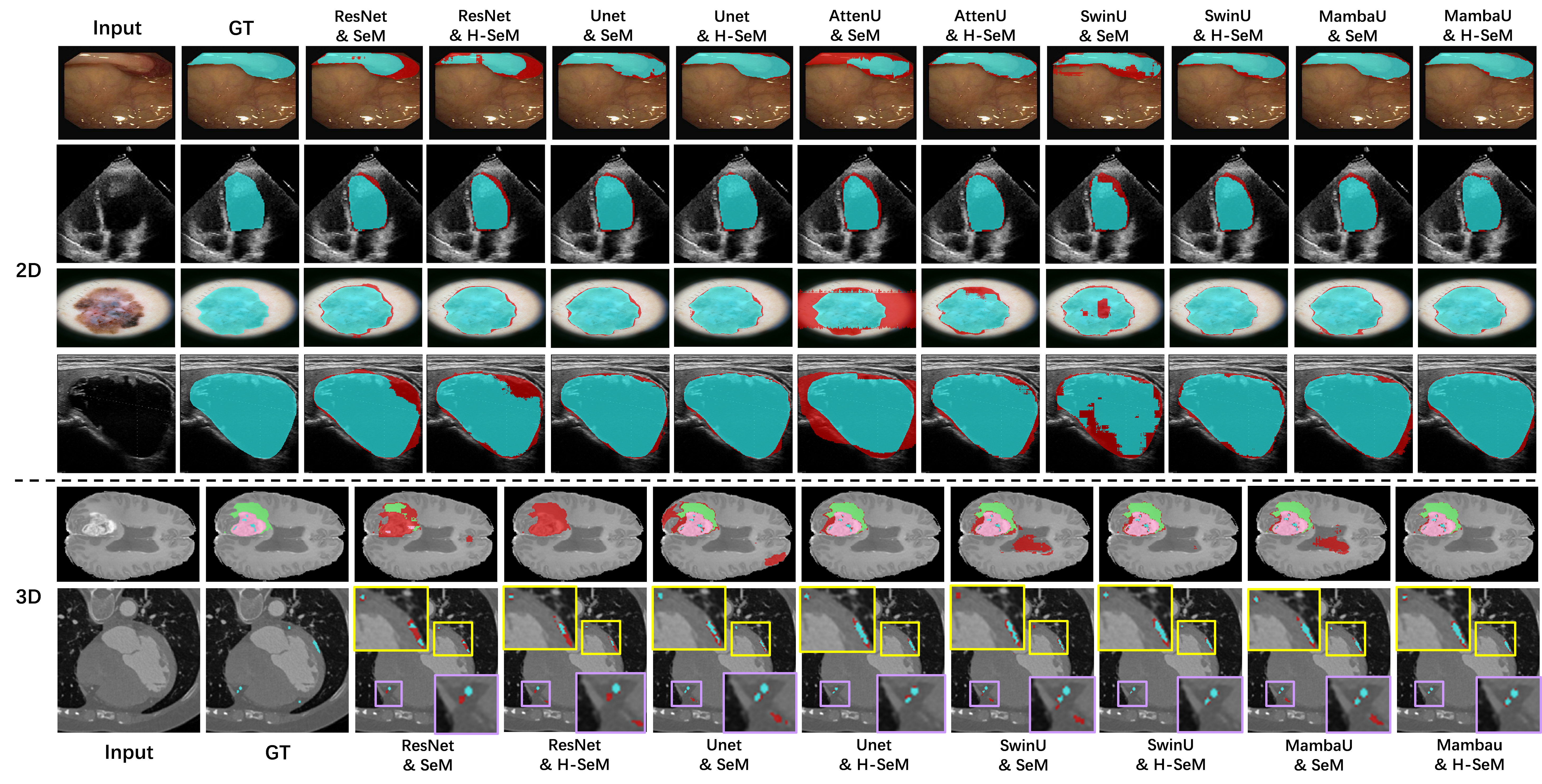
For segmentation, we evaluate our methods on six public datasets: CVC-ClinicDB, Echonet, ISIC, TN3K, BraTS21 (3D), ImageCAS (3D).
For reconstruction, we evaluate our methods on FastMRI.
Configuration files are in resources/img/biomed_2d and resources/img/biomed_3d.
Configuration file: resources/toy_ddpm.yaml
train_flemme --config resources/toy_ddpm.yaml
Configuration files are in resources/img/mnist
AutoEncoder & Variational AutoEncoder
Denoising Diffusion Probabilistic Model

Configuration files are in resources/img/cifar10
AutoEncoder & Variational AutoEncoder
Denoising Diffusion Probabilistic Model (conditional)

@misc{zhang2024flemmeflexiblemodularlearning,
title={Flemme: A Flexible and Modular Learning Platform for Medical Images},
author={Guoqing Zhang and Jingyun Yang and Yang Li},
year={2024},
eprint={2408.09369},
archivePrefix={arXiv},
primaryClass={eess.IV},
url={https://arxiv.org/abs/2408.09369},
}
Thanks to mamba, swin-transformer, diffusion model for their wonderful works.