GeneMap, A python tool for identifying the mapping of genes between two genomes by multi evidences.
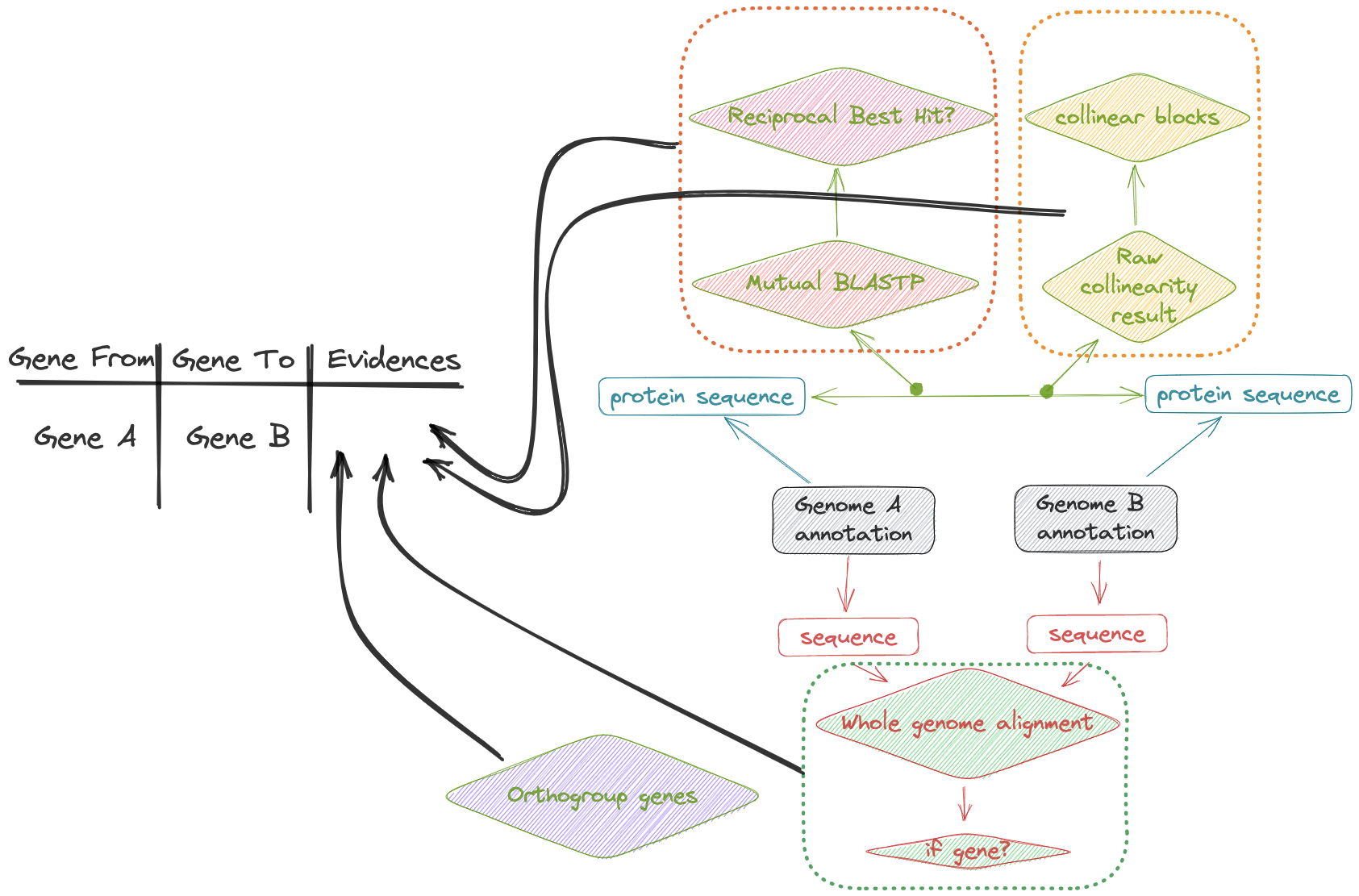
When we have multiple genomes of the same species (or related species), we usually need to identify the gene mapping relationship between the two genomes(Gene A in genome a, mapped to gene B in genome B). We used four alternative evidence, including:
-
RBH Reciprocal Best BLAST Hits
Despite these and other complication, the identification of reciprocal best hits for gene products is a good first approximation to the identification of orthologues in two or more organisms. It forms the basis for many orthology-finding tools, such as MCL, OrthoMCL and OrthoFinder. It can be carried out by carrying out BLAST+ searches using a short program, and this will be illustrated below.
-
ortholog OrthoFinder
-
synteny block McScanX
-
crosssmap minimap or AnchorWave
TODO
$ pip3 install GeneMap$ GeneMap.py -l list.txt -d work_dir -q GenomeA -t GenomeB -o output_prefixFeel free to dive in! Open an issue or submit PRs.
Standard Readme follows the Contributor Covenant Code of Conduct.
Thanks @songtaogui for design ideas.
GPL-3.0 © Weiwenjie