- Python > 3.6
- PyTorch=1.5.0
- FMRIB software library (FSL) 6.0
- Sundaresan V., Griffanti L., Jenkinson M. (2021) Brain Tumour Segmentation Using a Triplanar Ensemble of U-Nets on MR Images. In: Crimi A., Bakas S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science, vol 12658. Springer, Cham. [DOI: https://doi.org/10.1007/978-3-030-72084-1_31]
- Sundaresan, V., Griffanti, L. and Jenkinson, M., 2021. Brain tumour segmentation using a triplanar ensemble of U-Nets. arXiv preprint arXiv:2105.11356.
- Sundaresan, V., Zamboni, G., Rothwell, P.M., Jenkinson, M. and Griffanti, L., 2021. Triplanar ensemble U-Net model for white matter hyperintensities segmentation on MR images. Medical Image Analysis, p.102184. [DOI: https://doi.org/10.1016/j.media.2021.102184] (preprint available at [DOI: https://doi.org/10.1101/2020.07.24.219485])
Also cite the following paper if you use truenet fine-tuning:
- Sundaresan, V., Zamboni, G., Dinsdale, N. K., Rothwell, P. M., Griffanti, L., & Jenkinson, M. (2021). Comparison of domain adaptation techniques for white matter hyperintensity segmentation in brain MR images. Medical Image Analysis, p. 102215. [DOI: https://doi.org/10.1016/j.media.2021.102215] (preprint available at [DOI: https://doi.org/10.1101/2021.03.12.435171])
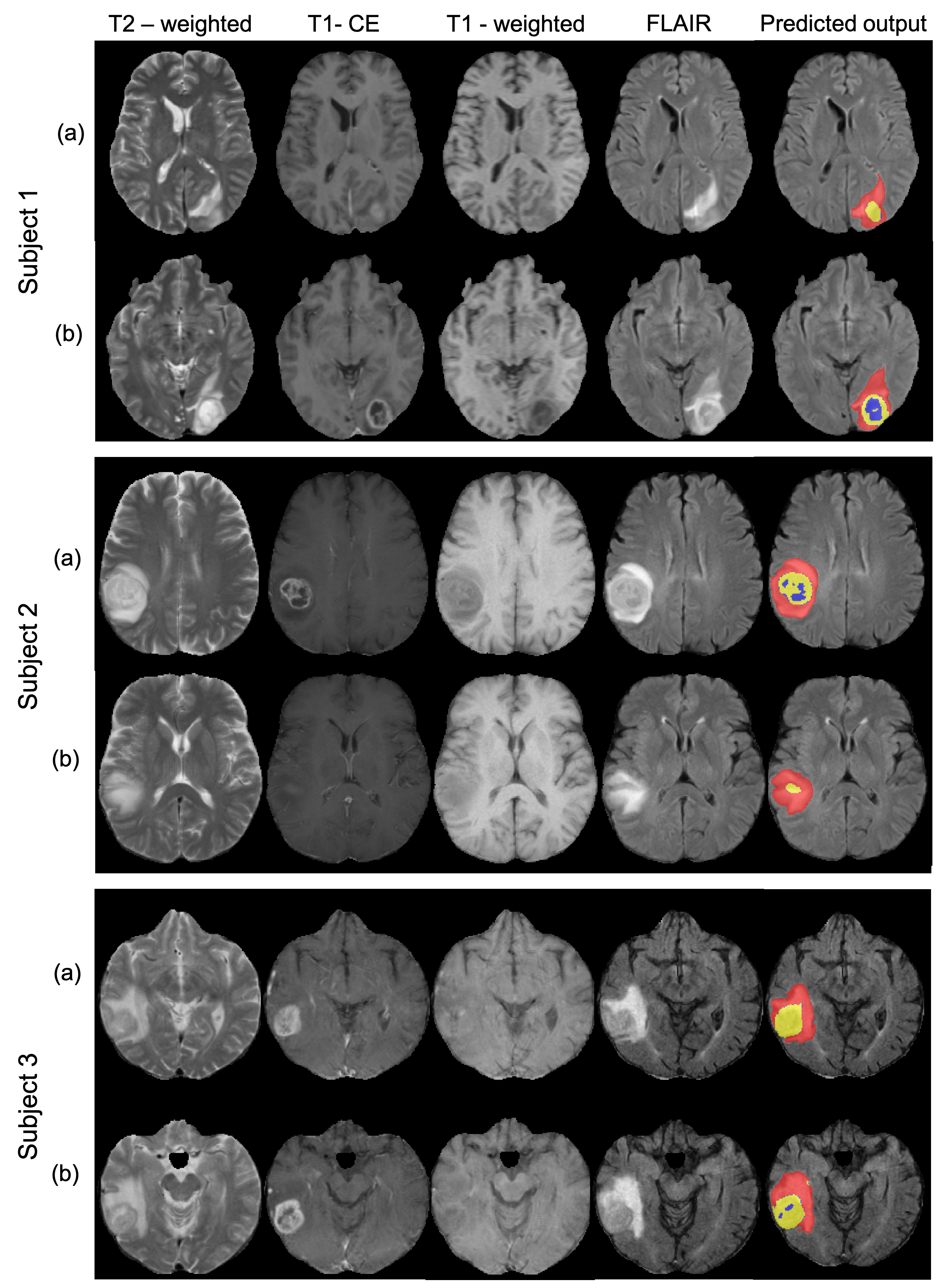
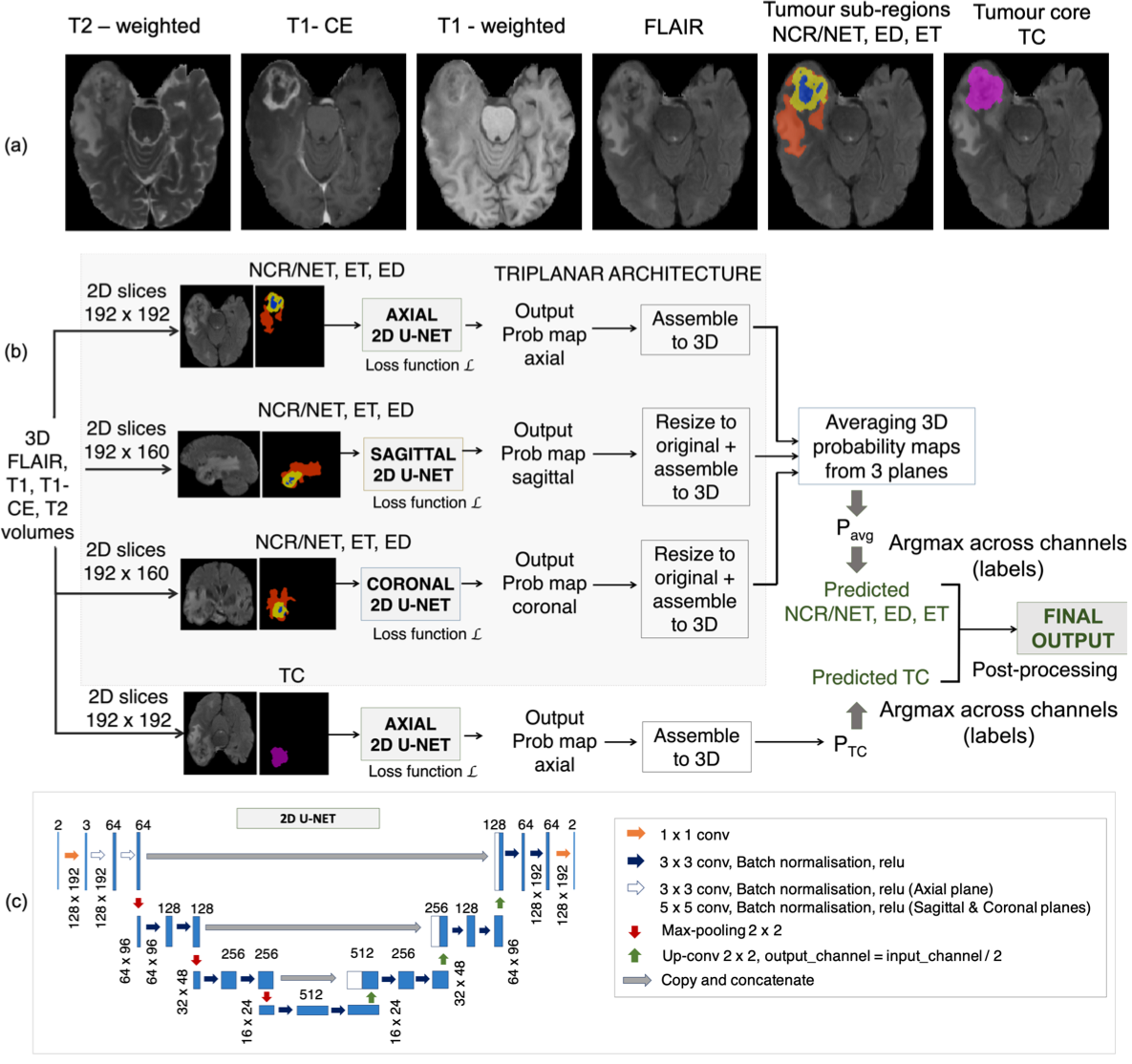
For each subject, the given input modalities include FLAIR, T1-weighted (T1), post-contrast T1-weighted (T1-CE) and T2-weighted (T2) images. The manual segmentations for the training dataset consists of 3 labels: necrotic core/non-enhancing tumour (NCR/NET), edematous tissue or peritumoral edema (ED) and enhancing tumour (ET). We used a sum of the voxel-wise cross-entropy loss function and the Dice loss as the total cost function.
Clone the git repository into your loacal directory and run:
python setup.py install
To find about the subcommands available in truenet:
truenet_tumseg --help
And for options and inputs for each sub-command, type:
truenet_tumseg <subcommand> --help (e.g. truenet train --help)
We used FLAIR, T1, T1 CE and T2 as inputs for the model. We reoriented the images to the standard MNI space, performed skull-stripping FSL BET and registered the T1-weighted image to the FLAIR using linear rigid-body registration.
Usage: prepare_tumseg_data --basemodality=<basemodality_name> --outname=<output_basename> --FLAIR=<FLAIR_image_name> --T1=<T1_image_name> --T1ce=<T1ce_image_name> --T2=<T2_image_name> [--keepintermediate] [-savelog]
The script applies the preprocessing pipeline on FLAIR, T1, T1ce and T2 to be used in FSL truenet_tumseg with a specified output basename
Base_modality_name and output_basename are mandatory inputs
Remaining inputs are optional, image corresponding to Base_modality_name must be provided.
The script applies the preprocessing pipeline on FLAIR, T1 and WM mask to be used in FSL truenet with a specified output basename
basemodality_name = name of the modality that the rest will be registered to (preferable ~1mm iso);
valid options: flair, t1, t1ce, t2
output_basename = absolute/relative path for the processed FLAIR and T1 images; output_basename_FLAIR.nii.gz, output_basename_T1.nii.gz etc. will be saved
FLAIR_image_name = absolute/relative path of the input unprocessed FLAIR image with the nifti file
T1_image_name = absolute/relative path of the input unprocessed T1 image with the nifti file
T1ce_image_name = name of the input unprocessed T1-contrast enhanced image
T2_image_name = name of the input unprocessed T2 image
specify --keepintermediate if you want to save intermediate results
specify --savelog for saving the comments (steps in preprocessing) in a log file
For example, if you have flair, t1 and t2 and want to register everything to t1, and want to keep intermediate files, use the following command
prepare_tumseg_data --basemodality=t1 --outname=path/to/outputbasename --FLAIR=path/to/input_flair.nii.gz --T1=path/to/input_t1.nii.gz --T2=path/to/input_t2.nii.gz --keepintermediate
Triplanar ensemble U-Net model, v1.0.1
Subcommands available:
- truenet_tumseg train Training a TrUE-Net model from scratch
- truenet_tumseg evaluate Applying a saved/pretrained TrUE-Net model for testing
- truenet_tumseg fine_tune Fine-tuning a saved/pretrained TrUE-Net model from scratch
- truenet_tumseg cross_validate Leave-one-out validation of TrUE-Net model
Usage: truenet_tumseg train -i <input_directory> -l <label_directory> -m <model_directory> [options]
Compulsory arguments:
-i, --inp_dir Path to the directory containing FLAIR and T1 images for training
-l, --label_dir Path to the directory containing manual labels for training
-m, --model_dir Path to the directory where the training model or weights need to be saved
Optional arguments:
-modality, --select_modality Input modalities available to train; format={FLAIR, T1, T1ce, T2, Others} (default=1 1 1 1 0)
-tr_prop, --train_prop Proportion of data used for training [0, 1]. The rest will be used for validation [default = 0.8]
-bfactor, --batch_factor Number of subjects to be considered for each mini-epoch [default = 10]
-loss, --loss_function Applying spatial weights to loss function. Options: weighted, nweighted [default=weighted]
-gdir, --gmdist_dir Directory containing GM distance map images. Required if -loss=weighted [default = None]
-vdir, --ventdist_dir Directory containing ventricle distance map images. Required if -loss=weighted [default = None]
-nclass, --num_classes Number of classes to consider in the target labels (nclass=2 will consider only 0 and 1 in labels;
any additional class will be considered part of background class [default = 2]
-plane, --acq_plane The plane in which the model needs to be trained. Options: axial, sagittal, coronal, all [default = all]
-da, --data_augmentation Applying data augmentation [default = True]
-af, --aug_factor Data inflation factor for augmentation [default = 2]
-sv_resume, --save_resume_training Whether to save and resume training in case of interruptions (default-False)
-ilr, --init_learng_rate Initial LR to use in scheduler [0, 0.1] [default=0.001]
-lrm, --lr_sch_mlstone Milestones for LR scheduler (e.g. -lrm 5 10 - to reduce LR at 5th and 10th epochs) [default = 10]
-gamma, --lr_sch_gamma Factor by which the LR needs to be reduced in the LR scheduler [default = 0.1]
-opt, --optimizer Optimizer used for training. Options:adam, sgd [default = adam]
-eps, --epsilon Epsilon for adam optimiser (default=1e-4)
-mom, --momentum Momentum for sgd optimiser (default=0.9)
-bs, --batch_size Batch size used for training [default = 8]
-ep, --num_epochs Number of epochs for training [default = 60]
-es, --early_stop_val Number of epochs to wait for progress (early stopping) [default = 20]
-sv_mod, --save_full_model Saving the whole model instead of weights alone [default = False]
-cv_type, --cp_save_type Checkpoint to be saved. Options: best, last, everyN [default = last]
-cp_n, --cp_everyn_N If -cv_type=everyN, the N value [default = 10]
-v, --verbose Display debug messages [default = False]
-h, --help. Print help message
Usage: truenet_tumseg evaluate -i <input_directory> -m <model_directory> -o <output_directory> [options]
Compulsory arguments:
-i, --inp_dir Path to the directory containing FLAIR and T1 images for testing
-m, --model_name Model basename with absolute path (will not be considered if optional argument -p=True)
-o, --output_dir Path to the directory for saving output predictions
Optional arguments:
-modality, --select_modality Input modalities available to train; format={FLAIR, T1, T1ce, T2, Others} (default=1 1 1 1 0)
-p, --pretrained_model Whether to use a pre-trained model, if selected True, -m (compulsory argument will not be onsidered) [default = False]. The model was pretrained on MICCAI BraTS 2020 Training dataset on 4 modalities: FLAIR, T1, T1ce, T2.
-nclass, --num_classes Number of classes in the labels used for training the model (for both pretrained models, -nclass=2) default = 2]
-post, --postprocessing Whether to perform post-processing for TC region (default=True)
-int, --intermediate Saving intermediate prediction results (individual planes) for each subject [default = False]
-cv_type, --cp_load_type Checkpoint to be loaded. Options: best, last, everyN [default = last]
-cp_n, --cp_everyn_N If -cv_type = everyN, the N value [default = 10]
-v, --verbose Display debug messages [default = False]
-h, --help. Print help message
Usage: truenet_tumseg fine_tune -i <input_directory> -l <label_directory> -m <model_directory> -o <output_directory> [options]
Compulsory arguments:
-i, --inp_dir Path to the directory containing FLAIR and T1 images for fine-tuning
-l, --label_dir Path to the directory containing manual labels for training
-m, --model_dir Path to the directory where the trained model/weights were saved
-o, --output_dir Path to the directory where the fine-tuned model/weights need to be saved
Optional arguments:
-modality, --select_modality Input modalities available to train; format={FLAIR, T1, T1ce, T2, Others} (default=1 1 1 1 0)
-p, --pretrained_model Whether to use a pre-trained model, if selected True, -m (compulsory argument will not be considered) [default = False]
-pmodel, --pretrained_model_name Pre-trained model to be used: mwsc, ukbb [default = mwsc]
-cpld_type, --cp_load_type Checkpoint to be loaded. Options: best, last, everyN [default = last]
-cpld_n, --cpload_everyn_N If everyN option was chosen for loading a checkpoint, the N value [default = 10]
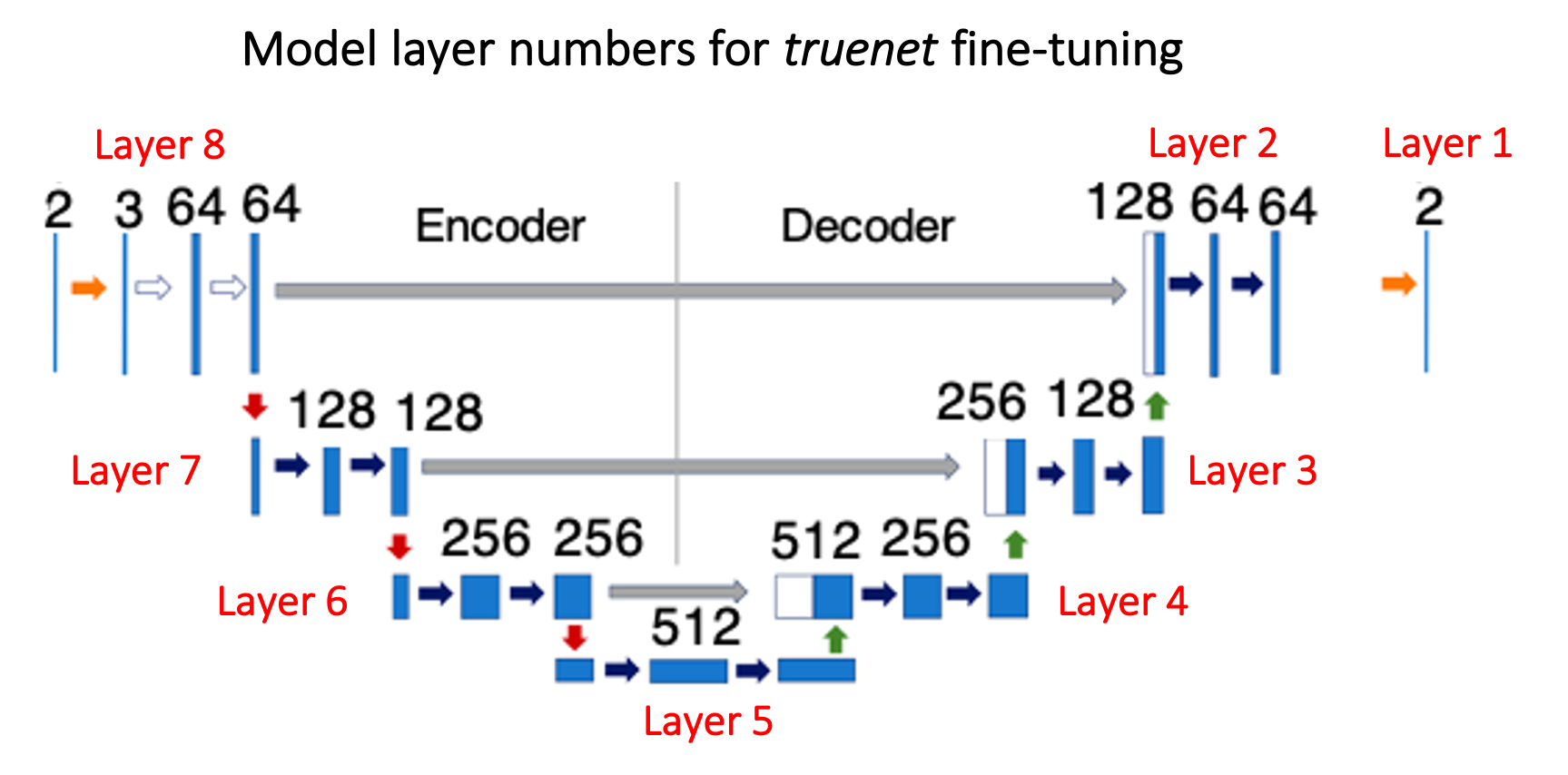
-ftlayers, --ft_layers Layers to fine-tune starting from the decoder (e.g. 1 2 -> final two two decoder layers, refer to the figure above)
-tr_prop, --train_prop Proportion of data used for fine-tuning [0, 1]. The rest will be used for validation [default = 0.8]
-bfactor, --batch_factor Number of subjects to be considered for each mini-epoch [default = 10]
-loss, --loss_function Applying spatial weights to loss function. Options: weighted, nweighted [default=weighted]
-gdir, --gmdist_dir Directory containing GM distance map images. Required if -loss = weighted [default = None]
-vdir, --ventdist_dir Directory containing ventricle distance map images. Required if -loss = weighted [default = None]
-nclass, --num_classes Number of classes to consider in the target labels (nclass=2 will consider only 0 and 1 in labels;
any additional class will be considered part of background class [default = 2]
-plane, --acq_plane The plane in which the model needs to be fine-tuned. Options: axial, sagittal, coronal, all [default all]
-da, --data_augmentation Applying data augmentation [default = True]
-af, --aug_factor Data inflation factor for augmentation [default = 2]
-sv_resume, --save_resume_training Whether to save and resume training in case of interruptions (default-False)
-ilr, --init_learng_rate Initial LR to use in scheduler for fine-tuning [0, 0.1] [default=0.0001]
-lrm, --lr_sch_mlstone Milestones for LR scheduler (e.g. -lrm 5 10 - to reduce LR at 5th and 10th epochs) [default = 10]
-gamma, --lr_sch_gamma Factor by which the LR needs to be reduced in the LR scheduler [default = 0.1]
-opt, --optimizer Optimizer used for fine-tuning. Options:adam, sgd [default = adam]
-eps, --epsilon Epsilon for adam optimiser (default=1e-4)
-mom, --momentum Momentum for sgd optimiser (default=0.9)
-bs, --batch_size Batch size used for fine-tuning [default = 8]
-ep, --num_epochs Number of epochs for fine-tuning [default = 60]
-es, --early_stop_val Number of fine-tuning epochs to wait for progress (early stopping) [default = 20]
-sv_mod, --save_full_model Saving the whole fine-tuned model instead of weights alone [default = False]
-cv_type, --cp_save_type Checkpoint to be saved. Options: best, last, everyN [default = last]
-cp_n, --cp_everyn_N If -cv_type = everyN, the N value [default = 10]
-v, --verbose Display debug messages [default = False]
-h, --help. Print help message
Usage: truenet_tumseg cross_validate -i <input_directory> -l <label_directory> -o <output_directory> [options]
Compulsory arguments:
-i, --inp_dir Path to the directory containing FLAIR and T1 images for fine-tuning
-l, --label_dir Path to the directory containing manual labels for training
-o, --output_dir Path to the directory for saving output predictions
Optional arguments:
-modality, --select_modality Input modalities available to train; format={FLAIR, T1, T1ce, T2, Others} (default=1 1 1 1 0)
-fold, --cv_fold Number of folds for cross-validation (default = 5)
-resume_fold, --resume_from_fold Resume cross-validation from the specified fold (default = 1)
-tr_prop, --train_prop Proportion of data used for training [0, 1]. The rest will be used for validation [default = 0.8]
-bfactor, --batch_factor Number of subjects to be considered for each mini-epoch [default = 10]
-loss, --loss_function Applying spatial weights to loss function. Options: weighted, nweighted [default=weighted]
-gdir, --gmdist_dir Directory containing GM distance map images. Required if -loss = weighted [default = None]
-vdir, --ventdist_dir Directory containing ventricle distance map images. Required if -loss = weighted [default = None]
-nclass, --num_classes Number of classes to consider in the target labels (nclass=2 will consider only 0 and 1 in labels;
any additional class will be considered part of background class [default = 2]
-plane, --acq_plane The plane in which the model needs to be trained. Options: axial, sagittal, coronal, all [default = all]
-da, --data_augmentation Applying data augmentation [default = True]
-af, --aug_factor Data inflation factor for augmentation [default = 2]
-sv_resume, --save_resume_training Whether to save and resume training in case of interruptions (default-False)
-ilr, --init_learng_rate Initial LR to use in scheduler for training [0, 0.1] [default=0.0001]
-lrm, --lr_sch_mlstone Milestones for LR scheduler (e.g. -lrm 5 10 - to reduce LR at 5th and 10th epochs) [default = 10]
-gamma, --lr_sch_gamma Factor by which the LR needs to be reduced in the LR scheduler [default = 0.1]
-opt, --optimizer Optimizer used for training. Options:adam, sgd [default = adam]
-eps, --epsilon Epsilon for adam optimiser (default=1e-4)
-mom, --momentum Momentum for sgd optimiser (default=0.9)
-bs, --batch_size Batch size used for fine-tuning [default = 8]
-ep, --num_epochs Number of epochs for fine-tuning [default = 60]
-es, --early_stop_val Number of fine-tuning epochs to wait for progress (early stopping) [default = 20]
-int, --intermediate Saving intermediate prediction results (individual planes) for each subject [default = False]
-v, --verbose Display debug messages [default = False]
-h, --help. Print help message
If you use the tool from this repository, please cite the following papers (journal publications to be updated, currently under review):
- Sundaresan V., Griffanti L., Jenkinson M. (2021) Brain Tumour Segmentation Using a Triplanar Ensemble of U-Nets on MR Images. In: Crimi A., Bakas S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science, vol 12658. Springer, Cham. [DOI: https://doi.org/10.1007/978-3-030-72084-1_31]
- Sundaresan, V., Griffanti, L. and Jenkinson, M., 2021. Brain tumour segmentation using a triplanar ensemble of U-Nets. arXiv preprint arXiv:2105.11356.
- Sundaresan, V., Zamboni, G., Rothwell, P.M., Jenkinson, M. and Griffanti, L., 2021. Triplanar ensemble U-Net model for white matter hyperintensities segmentation on MR images. Medical Image Analysis, p.102184. [DOI: https://doi.org/10.1016/j.media.2021.102184] (preprint available at [DOI: https://doi.org/10.1101/2020.07.24.219485]).
- Sundaresan, V., Zamboni, G., Dinsdale, N. K., Rothwell, P. M., Griffanti, L., & Jenkinson, M. (2021). Comparison of domain adaptation techniques for white matter hyperintensity segmentation in brain MR images. Medical Image Analysis, p. 102215. [DOI: https://doi.org/10.1016/j.media.2021.102215] (preprint available at [DOI: https://doi.org/10.1101/2021.03.12.435171]).