A prototype application that ranks species for conservation and research based on their conversation value, demand for samples by the community, and the samples already in biobank storage. This priority system assists biobanks with their resource allocation by prioritising species for resources, enabling them to enhance the diversity and availability of zoological samples for conservation and research. The application is in ongoing development and may therefore throw up bugs and unexpected errors.
- About the project
- Installation Guide
- Getting started
- Project Structure
- Note to developers
- Acknowledgements
- License
To get started with the app, read the installation guide. Once you've installed the app, you can run it from the CMD with python app.py.
- Windows
- Mac
- Linux
Once the application has been installed and app.py is running, users can navigate to the website by clicking the link to the host from CMD. The website has a landing page called 'home' which provides a brief overview of the project. To see other pages such as 'dataset' and 'upload' page, go to the navigation bar at the top of the page and click on the page links.
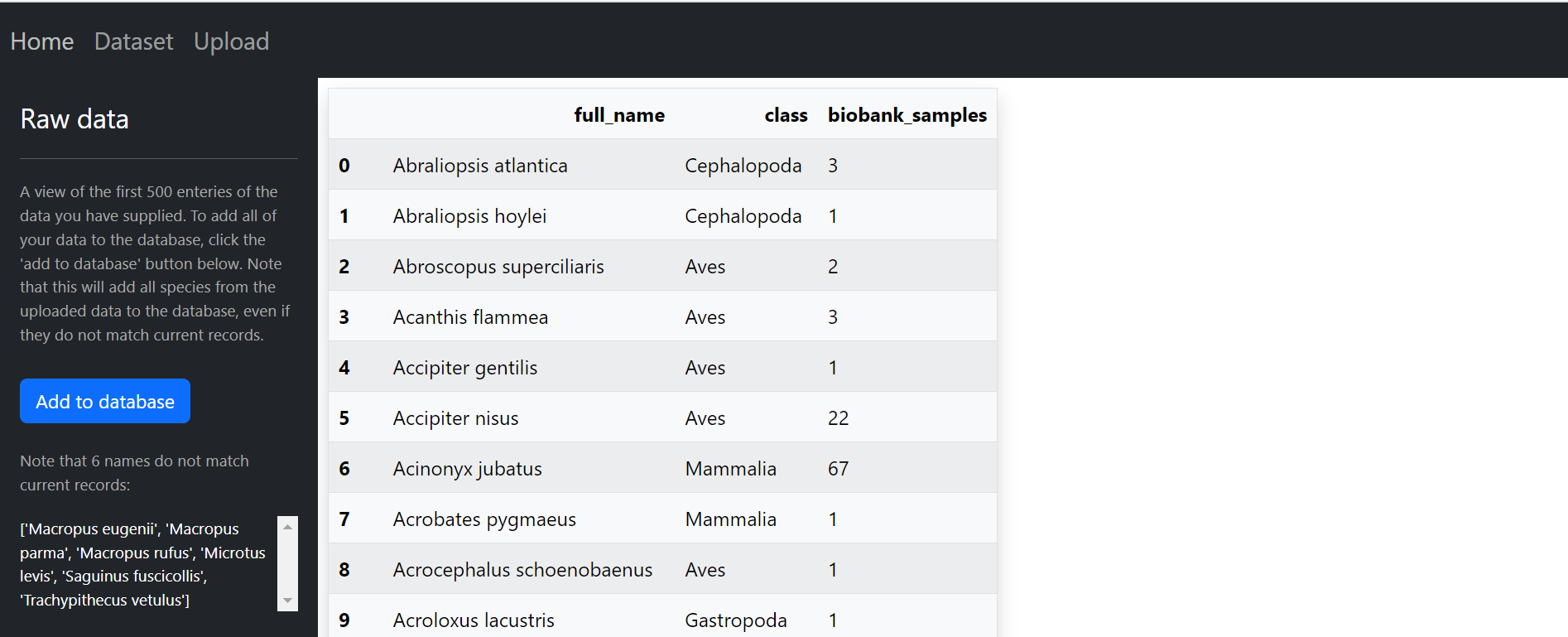
When you first click on the 'dataset' page is will re-direct you to the upload page, here you can upload the CryoArks (biobank sample) data located in the /datasets folder of this app. The added sample data will be displayed alongside the other categories in the dataset page, and it will contribute to the priority score calculated for each species. In the image below, you can see the CryoArks data being displayed in the upload page.
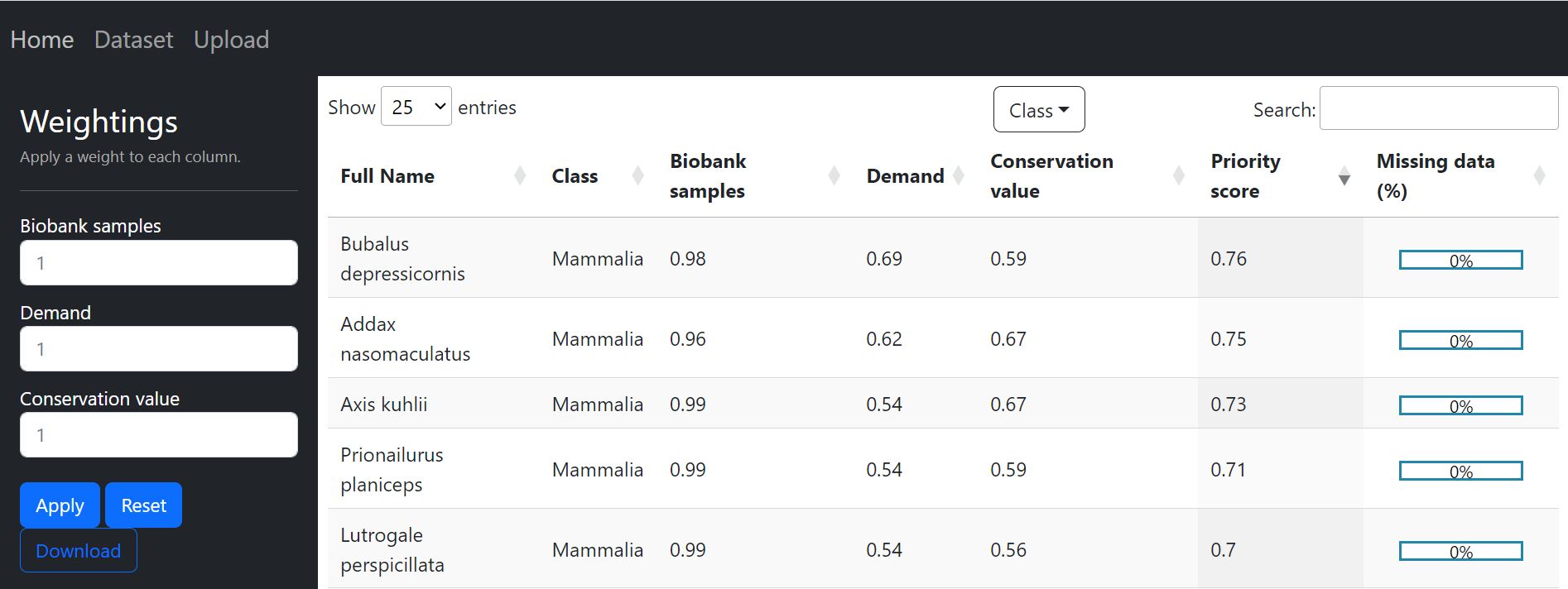
The dataset page displays scores for each category: Conservation Value (CV), Demand, and Biobank Samples. In addition, the class of each species, percentage of missing data (from the CV score) and the priority score derived from the category scores are displayed for each species. You can download the displayed data in the dataset page using the download button, all downloads can be found in the /downloaded folder inside the project folder. If you have trouble locates files in this project, then have a look at the Project Structure.
This project is still under development and may have some teething issues. If you run into a bug, please let me know and I will look into fixing it.
If you are interested in contributing to this project then please contact the author of the tool.
This project is in ongoing development. The previous version was developed on the request to have the data in SQL for frequent access and use in other projects. This current version allows user to specify if they want to use an optional SQL database to store the data.
There are some areas I am planning to work on in future editions:
- Replacing fake demand scores with real demand/ request data
- Improving error messaging for more precise error handling
- Handling missing data effectively so it doesn't cause artificially low scores
- Enforcing more rigorous data health checks to avoid unexpected errors/ bugs
Mike Bruford inspired and directed this project, he provided the methodology implemented here. Mike was the director of The Frozen Ark Project and the lead investigator of CryoArks. He sadly passed away in April 2023.
Mafalda Costa provided ongoing support for the development of this project. Maf is a conservation biologist at Cardiff University and a research associate for CryoArks.
Thank you to Matthew Grainger for providing information on the MAPISCo methodology that this project adopts.
Thank your to IUCN, CITES, EDGE and CryoArks for your data.
IUCN 2022. IUCN Red List of Threatened Species. Version 2022-2 The EDGE2 protocol: Advancing the prioritisation of Evolutionarily Distinct and Globally Endangered species for practical conservation action Gumbs R, Gray CL, Böhm M, Burfield IJ, Couchman OR, et al. (2023) PLOS Biology 21(2): e3001991. UNEP (2023). The Species+ Website. Nairobi, Kenya. Compiled by UNEP-WCMC, Cambridge, UK. CryoArks (2019). CryoArks Database. Method for the Assesment of Priorities for International Species Conservation (MAPISCo, Defra)This tool was built soley by Sarah Harwood @sjharw
This app is published under a General Public License (GPL) v2.0. Please reference the original author of this tool @sjharw in any derivative/ modified/ copied versions of this work.
The goal of the GPL v2.0 is to promote the free sharing and collaboration of software. It ensures that everyone has the freedom to use, modify, and share software while respecting the rights of others.
The GPL grants you the following freedoms:
- You can use the software without paying or needing permission
- You can modify the software to suit you needs, but you must share your modifications under GPL v2.0
- You can share the software, but you must include the GPL v2.0 license along with any distributions
- You cannot prevent others from using, modifying, or sharing the software as the license permits