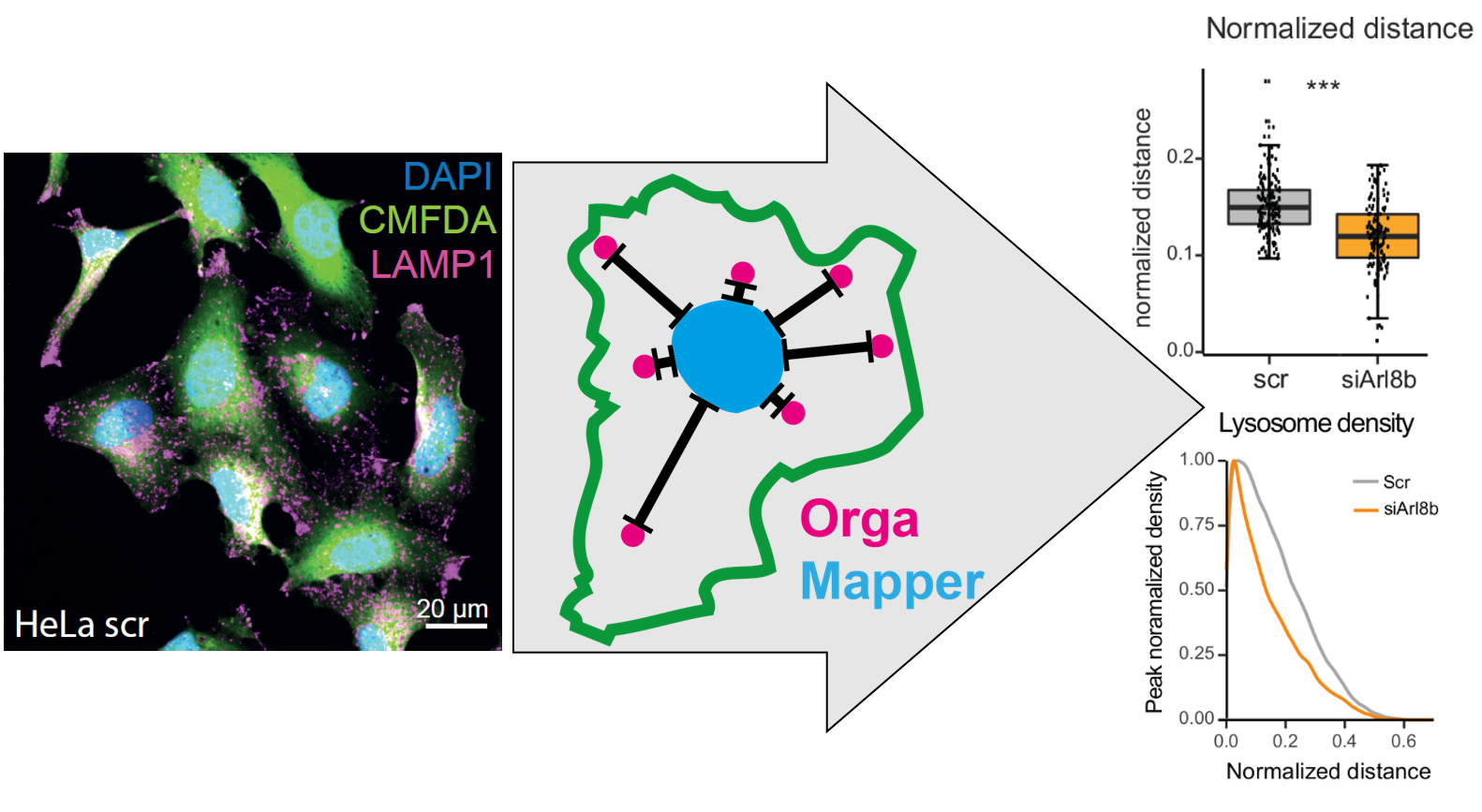
Easy to use plugin for analysing organelle position
The aim of the OrgaMapper workflow is to measure and map organelle distribution within cells with ease. The distance of the organelles are related to the nucleus using location or signal intensity.
Documentation
Have a look at the github pages site for more information:
https://schmiedc.github.io/OrgaMapper/
Introduction
The image analysis plugin solves 3 core image analysis tasks:
1. Nucleus segmentation: The nucleus is segmented using an intensity threshold. Nuclei at the edge of the field of view are rejected. The generated masks are filtered for size and shape.
2. Cell segmentation: Cells are segmented using an intensity threshold. Touching cells are separated using a marker controlled watershed. The cell ROIs are filtered such that each cell contains 1 nucleus. Cells are further filered by size and shape.
3. Organelle detection: We use a blob detection to detect individual organelles to locate their number and the position within the cell.
The generated masks and ROIs are then used to perform the following measurements:
- Filtered cell ROIs:
- Intensity of the organelle channel and an optional measurement channel.
- Ferets diameter as well as the cell area.
- Filtered cell ROIs & Nucleus Mask:
- Compute euclidean distance map (EDM) from edge of nucleus masks.
- Measure distance of each organelle detection based on EDM.
- Extract signal value and distance of the organelle channel and an optional measurement channel.
- Outside of unfilte1. red cell ROIs:
- Background of the organelle channel and an optional measurement channel.
Accepted datasets
Single multichannel .tiff files and multichannel multiseries files. We tested the workflow on multiseries .nd2 files from Nikon CSU.
For the Data analysis and plotting to work seamlessly with the image analysis we require the following naming pattern.
Single .tif files:
<Name>_<Treatment>_<Number>
Multiseries files:
<Name>_<Treatment/Wellnumber>
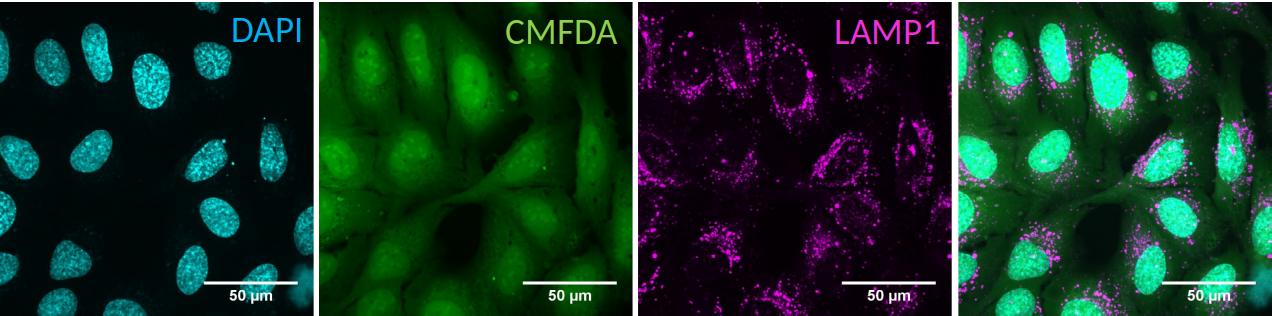
The data is expected to contain a channel with nucleus staining (DAPI) staining against cytoplasm (CMFDA) and against an organelle of choice.
Installation
For the image analysis you need to download and install Fiji: https://fiji.sc/ The plugin is available via an update site. Add the Cellular-Imaging site:
- Select Help › Update... from the menu bar.
- Click on Manage update sites. Which opens the Manage update sites dialog.
- Press Add update size a new line in the Manage update sites dialog appears
- Add https://sites.imagej.net/Cellular-Imaging/ as url
- Add an optional name such as Cellular-Imaging
- Press Close and then Apply changes
For the data anaylsis you need to install R: https://www.r-project.org/
As an R editor I recommend to use RStudio: https://rstudio.com/products/rstudio/download/