Table of contents
- Description
- How it works
- Getting started with App
- CLI
- login
- create
- deploy
- apps
- Roadmap
- Citations
- License
DeepChain apps is a collaborative framework that allows the user to create scorers to evaluate protein sequences. These scorers can be either classifier or predictor.
This github is hosting a template for creating a personal application to deploy on deepchain.bio. The main deepchain-apps package can be found on pypi. To leverage the apps capability, take a look at the bio-transformers and bio-datasets package.
It is recommended to work with conda environnements in order to manage the specific dependencies of the package.
conda create --name deepchain-env python=3.7 -y
conda activate deepchain-env
pip install deepchain-appsIf you want to create and deploy an app on deepchain hub, you could use the command provided in the deepchain-apps package. Below are the main commands that should be used in a terminal:
deepchain login
deepchain create myapplication
The last command will download the github files inside the myapplication folder.
You can modify the app.py file, as explained in the Deepchain-apps templates
To deploy the app on deepchain.bio, use:
deepchain deploy myapplication
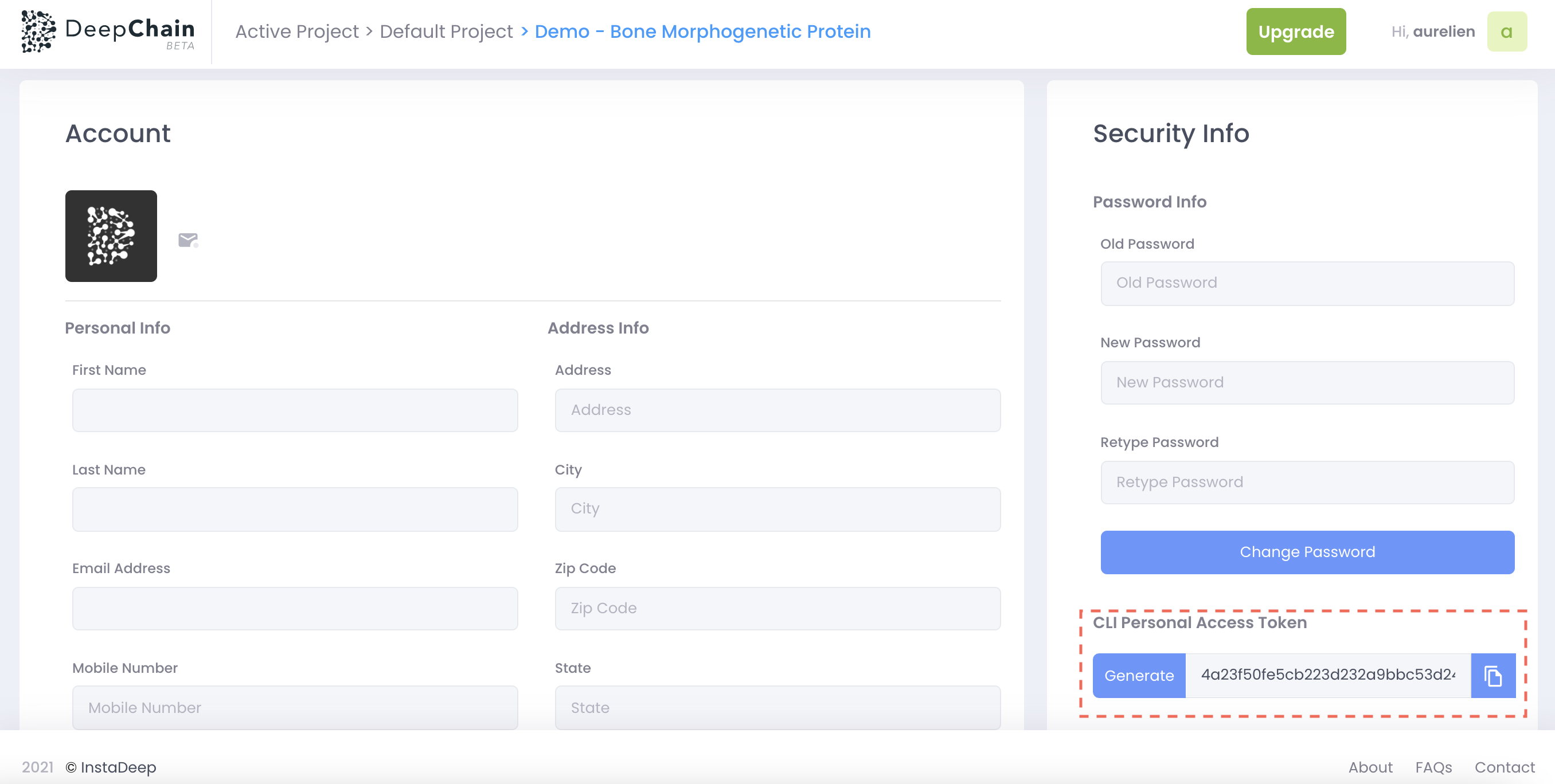
If you want to deploy biology app on deepchain, you should first create a personal account on deepchain and go to the user profile section. As you can see below, you will be able to generate a PAT (personal access token) that you can use with the CLI command:
deepchain login
When creating an app, you will download the current github folder with the following structure.
.
├── README.md # explains how to create an app
├── __init__.py # __init__ file to create python module
├── checkpoint
│ ├── __init__.py
│ └── Optionnal : model.pt # optional: model to be used in app must be placed there
├── examples
│ ├── app_with_checkpoint.py # example: app example with checkpoint
│ └── torch_classifier.py # example: show how to train a neural network with pre-trained embeddings
└── src
├── DESC.md # Desciption file of the application, feel free to put a maximum of information.
├── __init__.py
├── app.py # main application script. Main class must be named App.
└── Optional : model.py # file to register the models you use in app.py.
└── tags.json # file to register the tags on the hub.
The main class must be named App in app.py
For your app to be visible and well documented, tags should be filled to precise at least the tasks section. It will be really useful to retrieve it from deepchain hub.
- tasks
- librairies
- embeddings
- datasets
- device
If you want your app to benefit from deepchain' GPU, set device to "gpu" in tags. It will run on "cpu" by default.
DeepChainApp class provides two special methods to load checkpoint and extra files:
get_checkpoint_path(__file__): return path for file in checkpoint folderget_filepath(__file__,file): return path for file in src folder
You must use these functions, not a relative path to load your extra files in order to safely load your scorer in deepchain.
You can also create an application based on an app already available on the public deepchain hub:
First, you can list all the available app in the hub like following:
>> deepchain apps --public
----------------------------------------------------------------
APP USERNAME
----------------------------------------------------------------
OntologyPredict username1@instadeep.com
DiseaseRiskApp username2@instadeep.com
You can simply download the app locally with the cli:
deepchain download username1@instadeep.com/OntologyPredict OntologyPredict
The app will be downloaded in the OntologyPredict folder.
Some templates are provided in order to create and deploy an app.
You can implement whatever function you want inside compute_scores() function.
It just has to respect the return format:
One dictionary for each protein that is scored. Each key of the dictionary are declared in score_names() function.
[
{
'score_names_1':score11
'score_names_2':score21
},
{
'score_names_1':score12
'score_names_2':score22
}
]An example of training with an embedding is provided in the example/torch_classifier.py script.
Be careful, you must use the same embedding for the training and the compute_scores() method.
When training a model with pytorch, you must save the weights with the state_dict() method, rebuilt the model architecture in the Scorer or in a model.py file and load the weights like in the example below.
from typing import Dict, List, Optional
import torch
from biotransformers import BioTransformers
from deepchain.components import DeepChainApp
# TODO : from model import myModel
from deepchain.models import MLP
from torch import load
Score = Dict[str, float]
ScoreList = List[Score]
class App(DeepChainApp):
"""DeepChain App template:
- Implement score_names() and compute_score() methods.
- Choose a a transformer available on BioTranfformers
- Choose a personal keras/tensorflow model
"""
def __init__(self, device: str = "cuda:0"):
self._device = device
self.transformer = BioTransformers(backend="protbert", device=device)
# Make sure to put your checkpoint file in your_app/checkpoint folder
self._checkpoint_filename: Optional[str] = "model.pt"
# build your model
self.model = MLP(input_shape=1024, n_class=2)
# load_model for tensorflow/keras model-load for pytorch model
if self._checkpoint_filename is not None:
state_dict = load(self.get_checkpoint_path(__file__))
self.model.load_state_dict(state_dict)
self.model.eval()
@staticmethod
def score_names() -> List[str]:
"""App Score Names. Must be specified.
Example:
return ["max_probability", "min_probability"]
"""
return ["probability"]
def compute_scores(self, sequences: List[str]) -> ScoreList:
"""Return a list of all proteins score"""
x_embedding = self.transformer.compute_embeddings(sequences)["cls"]
probabilities = self.model(torch.tensor(x_embedding).float())
probabilities = probabilities.detach().cpu().numpy()
prob_list = [{self.score_names()[0]: prob[0]} for prob in probabilities]
return prob_list"""
Module that provide a classifier template to train a model on embeddings.
With use the pathogen vs human dataset as an example. The embedding of 100k proteins come
from the protBert model.
The model is built with pytorch_ligthning, a wrapper on top of
pytorch (similar to keras with tensorflow)
Feel feel to build you own model if you want to build a more complex one
"""
import numpy as np
from biodatasets import list_datasets, load_dataset
from deepchain.models import MLP
from deepchain.models.utils import confusion_matrix_plot, model_evaluation_accuracy
from sklearn.model_selection import train_test_split
# Load embedding and target dataset
pathogen = load_dataset("pathogen")
_, y = pathogen.to_npy_arrays(input_names=["sequence"], target_names=["class"])
embeddings = pathogen.get_embeddings("sequence", "protbert", "cls")
x_train, x_test, y_train, y_test = train_test_split(embeddings, y[0], test_size=0.3)
# Build a multi-layer-perceptron on top of embedding
# The fit method can handle all the arguments available in the
# 'trainer' class of pytorch lightening :
# https://pytorch-lightning.readthedocs.io/en/latest/common/trainer.html
# Example arguments:
# * specifies all GPUs regardless of its availability :
# Trainer(gpus=-1, auto_select_gpus=False, max_epochs=20)
# Input variables for MLP
n_class = len(np.unique(y_train))
input_shape = x_train.shape[1]
mlp = MLP(input_shape=input_shape, n_class=n_class)
mlp.fit(x_train, y_train, epochs=5)
mlp.save("model.pt")
# Model evaluation
y_pred = mlp(x_test).squeeze().detach().numpy()
model_evaluation_accuracy(y_test, y_pred)
# Plot confusion matrix
confusion_matrix_plot(y_test, (y_pred > 0.5).astype(int), ["0", "1"])The CLI provides 5 main commands:
-
login : you need to supply the token provide on the platform (PAT: personal access token).
deepchain login -
create : create a folder with a template app file
deepchain create my_application -
deploy : the code and checkpoint are deployed on the platform, you can select your app in the interface on the platform.
-
with checkpoint upload
deepchain deploy my_application --checkpoint -
Only the code
deepchain deploy my_application
-
-
apps :
-
Get info on all local/upload apps
deepchain apps --infos -
Remove all local apps (files & config):
deepchain apps --reset -
Remove a specific application (files & config):
deepchain apps --delete my_application -
List all public apps:
deepchain apps --public
-
-
download :
-
Download locally an app deployed on deepchain hub
deepchain download user.name@mail.com/AppName AppName
-
Apache License Version 2.0