| Branch |  |
 |
|---|---|---|
master |
||
develop |
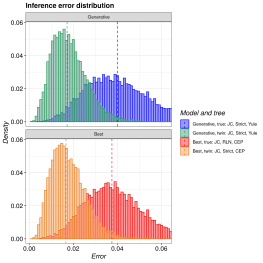
pirouette is an R package that estimates the error BEAST2 makes from a given
phylogeny. This phylogeny can be created using any (non-BEAST) speciation model,
for example the Protracted Birth-Death or Multiple-Birth-Death models.
nsm: Nucleotide Substitution Modeltral: TRue ALignmenttwal: TWin ALignment
See the FAQ.
See CONTRIBUTING, at Submitting use cases
See CONTRIBUTING, at 'Submitting code'
See CONTRIBUTING, at 'Submitting bugs'
Sure, just add an Issue. Or send an email.
| Package |  |
 |
|---|---|---|
| beautier | ||
| beastier | ||
| mauricer | ||
| mcbette | ||
| tracerer |
| Package |  |
 |
|---|---|---|
| beautier | ||
| beastier | ||
| mauricer | ||
| mcbette | ||
| tracerer |
| Package | Status |
|---|---|
| babette_on_windows | |
| beastier_on_windows | |
| beautier_on_windows | |
| mauricer_on_windows | |
| tracerer_on_windows |
- Bilderbeek, RJC, Laudanno, G, Etienne, RS. Quantifying the impact of an inference model in Bayesian phylogenetics. Methods Ecol Evol. 2020; 00: 1– 8. https://doi.org/10.1111/2041-210X.13514