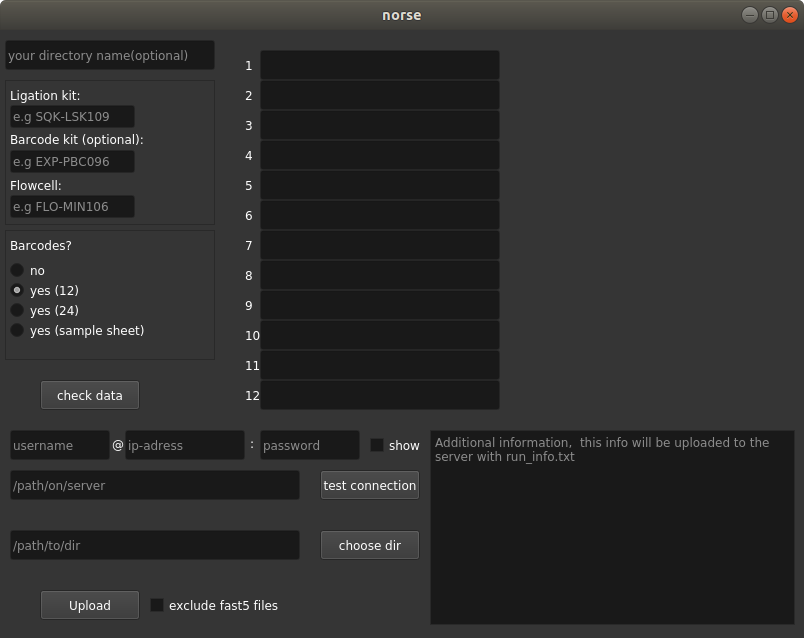
GUI for nanopore data transfer to a server and sample informations. All informations are collected in a run_info.txt file that is stored with the uploaded data.
- Install norse
- via conda
- via docker
- Usage
- conda installation here
# choose a suitable install location (e.g home dir)
git clone https://github.com/t3ddezz/norse.git
cd norse
# conda installation
conda env create -f environment.yml
conda activate norse
python setup.py install
-
Troubleshooting with this error: "This application failed to start because no Qt platform plugin could be initialized. Reinstalling the application may fix this problem. Available platform plugins are: eglfs, linuxfb,xcb."
- then try this:
sudo apt-get install libxkbcommon-x11-0
sudo apt install libxcb-xinerama0
- Check if the install worked
- Type (in the norse environment):
conda activate norse
norse -v
# clone git repository and navigate into the dir
git clone https://github.com/t3ddezz/norse.git && cd norse
# create docker image via
docker build -t norse:build .
# run docker image via (port mapping is missing here!!!)
docker run --rm \
-u qtuser \
-v /tmp/.X11-unix:/tmp/.X11-unix \
-e DISPLAY=unix$DISPLAY \
--network host \
-v $HOME:/home/qtuser \
-v $PWD:/upload \
norse:build \
norse -r
or use the newest docker from dockerhub:
# run docker image from dockerhub (insert respective VERSION)
docker run --rm \
-u qtuser \
-v /tmp/.X11-unix:/tmp/.X11-unix \
-e DISPLAY=unix$DISPLAY \
--network host \
-v $HOME:/home/qtuser \
-v $PWD:/upload \
dataspott/norse:VERSION \
norse -r
Run norse -r to start the program or norse -v to show the program version.