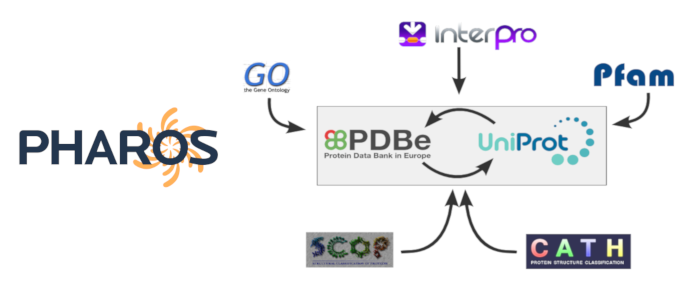
Here we use the Structure Integration with Function, Taxonomy and Sequence (SIFTS) database to map Pharos protein targets to PDB structures.
The goal is to obtain a dataset of human targets with both available structures and known ligand binding affinities. We also aim to obtain a characterizaton of the distribution of these PDB structures across different receptor families, such as Kinases, GPCRs, Ion Channels, Nuclear Receptors, and Transporters.
This repository contains datasets of Pharos targets in the Tchem and Tclin classes, and Jupyter notebooks for generating their PDB mappings.