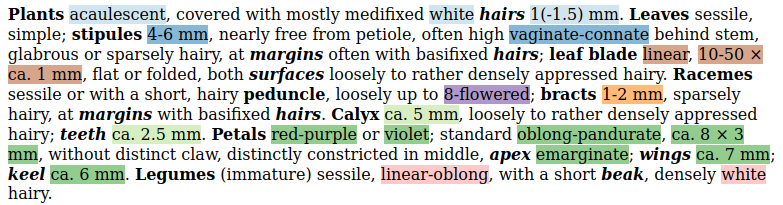
Challenge: Extract trait information from plant treatments. That is, if I'm given treatment text like: (Reformatted to emphasize targeted traits.)
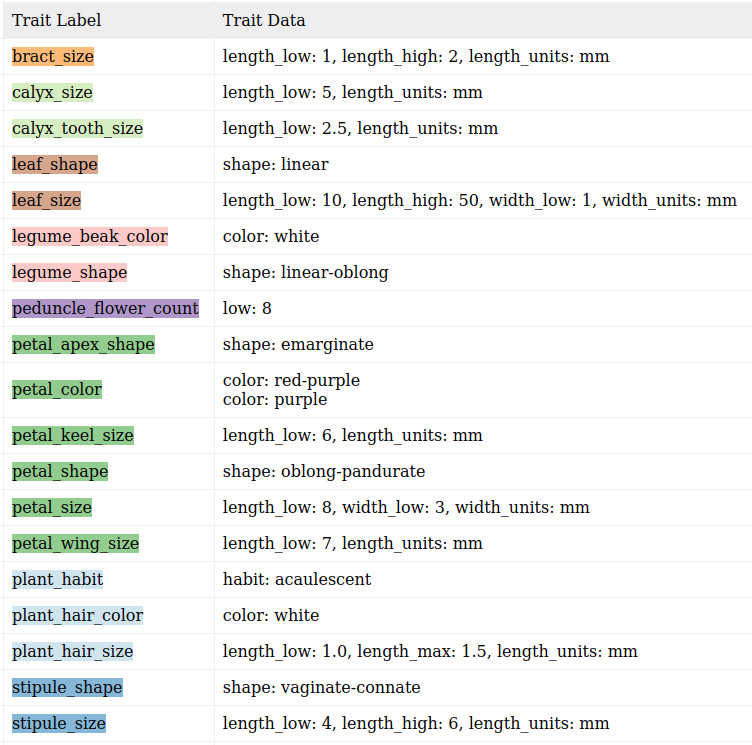
I should be able to extract: (Colors correspond to the text above.)
Essentially, we are finding relevant terms in the text (NER) and then linking them (Entity Linking). There are 5 types of terms:
- The traits themselves: These are things like color, size, shape, woodiness, etc. They are either a measurement, count, or a member of a controlled vocabulary.
- Plant parts: Things like leaves, branches, roots, seeds, etc. These have traits. So they must be linked to them.
- Plant subparts: Things like hairs, pores, margins, veins, etc. Leaves can have hairs and so can seeds. They also have traits and will be linked to them, but they must also be linked to a part to have any meaning.
- Sex: Plants exhibit sexual dimorphism, so we to note which part/subpart/trait notation is associated with which sex. 5Other text: Things like conjunctions, punctuation, etc. Although they are not recorded, they are often important for parsing and linking of terms.
- Rule based parsing. Most machine learning models require a substantial training dataset. I use this method to bootstrap the training data. If machine learning methods fail, I can fall back to this.
- Machine learning models. (In progress)
- I label terms using Spacy's phrase and rule-based matchers.
- Then I match terms using rule-based matchers repeatedly until I have built up a recognizable trait like: color, size, count, etc.
- Finally, I associate traits with plant parts.
For example, given the text: Petiole 1-2 cm.:
- I recognize vocabulary terms like:
Petioleis plant part1a number-a dash2a numbercmis a unit notation
- Then I group tokens. For instance:
1-2 cmis a range with units which becomes a size trait.
- Finally, I associate the size with the plant part
Petioleby using a tree base parser. Spacy will build a labeled sentence dependency tree. We look for patterns in the tree to link traits with plant parts.
There are, of course, complications and subtleties not outlined above, but you should get the gist of what is going on here.
You will need to have Python 3.11 (or later) installed. You can install the requirements into your python environment like so:
git clone https://github.com/rafelafrance/traiter_efloras.git
cd traiter_efloras
optional: virtualenv -p python3.11 .venv
optional: source .venv/bin/activate
make install
./extract.py ... TODO ...
Having a test suite is absolutely critical. The strategy I use is every new trait gets its own test set. Any time there is a parser error I add the parts that caused the error to the test suite and correct the parser. I.e. I use the standard red/green testing methodology.
You can run the tests like so:
cd /my/path/to/efloras_mimosa
python -m unittest discover