-- If you can get away with it, you may want to try an end-to-end machine learning approach.
I just spent a substantial effort on using a machine learning model, and I was wrong about the relative usefulness of rule-based and machine learning approaches for information extraction. Both have their strengths and weaknesses. I am now convinced that for the next couple of years you will need either a hybrid approach with, maybe, a custom model. However, I still do not recommend a rule only approach.
This is the rule-based Traiter information extraction/data mining library used by all client Traiter projects.
This contains only the rule-based parsers used for information extraction. The model-based parsers are in other repositories.
Some literature mined:
- PDFs containing research papers of descriptions of species.
- PDFs containing distribution data of species.
- Database downloads of field notes and species descriptions.
- Images of museum specimens. We are currently extracting data from the labels in the images.
- Data scraped from websites containing formal descriptions (treatments) of species.
- Images of PDFs of species descriptions and distribution data.
- Images of data collection notes.
Note All terms, traits, and extraction methods are unique to the body of literature being mined so this repository only contains truly universal terms, traits or, for that matter, functions used across many research domains.
- A trait is just an annotated spaCy entity. That is, I'm treating traits (aka attributes) as if they were their own entity. This has the advantage in that I can use named entity recognition (NER) techniques on them. We can also use standard entity link methods for linking traits to entities.
- There may (or not) be several layers of entities and traits. For instance, a plant SPECIES (a top-level entity) may have FLOWERs (a secondary or sub-entity) and those FLOWERs will have a set of characteristic COLORs (a trait).
- We leverage normal spaCy entities to build traits. Sometimes I will use spaCy's NER entities as building block traits.
- Have experts identify relevant terms and target traits.
- We use expert identified terms to label terms using spaCy's phrase matchers. These are sometimes traits themselves but are more often used as anchors for more complex patterns of traits.
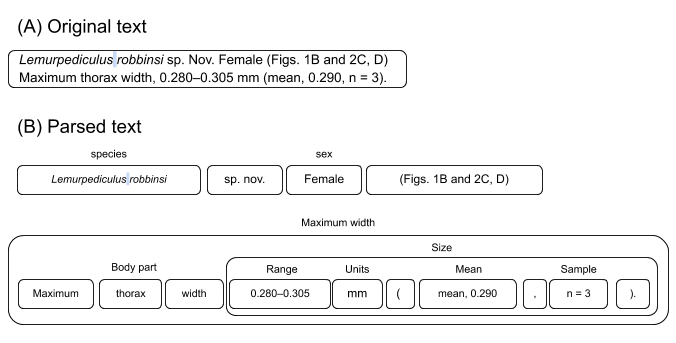
- We then build up more complex terms from simpler terms using spaCy's rule-based matchers repeatedly until there is a recognizable trait. See the image below.
- We may then link traits to each other (entity relationships) also using spaCy rules.
- Typically, a trait gets linked to a higher level entity like SPECIES <--- FLOWER <--- {COLOR, SIZE, etc.} and not peer to peer like PERSON <---> ORG.
- Also note that sometimes the highest level entity is assumed by its context. For instance, if a web page is a description of a newly found species then I don't need to parse the species name in the document.
You will need GIT to clone this repository. You will also need to have Python3.11+ installed, as well as pip, a package manager for Python. You can install the requirements into your python environment like so:
git clone https://github.com/rafelafrance/traiter.git
cd /path/to/traiter
make installThis repository is a library for other Traiter projects and is not designed to be run directly.
There are tests which you can run like so:
make test