AutoScore-Survival: Developing interpretable machine learning-based time-to-event scores with right-censored survival data
- AutoScore-Survival Introduction
- AutoScore-Survival
Demonstration
- Load R package
- Load data
- Data preprocessing (Users to check the following)
- AutoScore-Survival preprocessing (Users to check the following)
- **AutoScore-Survival Demo **
- Prepare training, validation, and test datasets
- STEP(i): Generate variable ranking list (AutoScore-Survival Module 1)
- STEP(ii): Select the best model with parsimony plot (AutoScore-Survival Modules 2+3+4)
- STEP(iii): Generate initial scores with the final list of variables (Re-run AutoScore-Survival Modules 2+3)
- STEP(iv): Fine-tune the initial score generated in STEP(iii) (AutoScore-Survival Module 5 & Re-run AutoScore-Survival Modules 2+3)
- STEP(v): Evaluate final risk scores on test dataset (AutoScore-Survival Module 6)
- AutoScore-Survival
AutoScore-Survival is a novel machine learning framework to automate the development of interpretable time-to-event scores. AutoScore-Survival consists of six modules: 1) variable ranking with machine learning, 2) variable transformation, 3) score derivation, 4) model selection, 5) domain knowledge-based score fine-tuning, and 6) performance evaluation. The AutoScore-Survival is elaborated in the article (http://dx.doi.org/10.2196/21798 and https://arxiv.org/abs/2106.06957). AutoScore-Survival could seamlessly generate risk scores based on survival data, which can be easily implemented and validated in clinical practice. Moreover, it enables users to build transparent and interpretable time-to-event scores quickly in a straightforward manner.
The five pipeline functions: AutoScore_Survival_rank(),
AutoScore_Survival_parsimony(), AutoScore_Survival_weighting(),
AutoScore_Survival_fine_tuning() and AutoScore_Survival_testing()
constitute the 5-step AutoScore-based process for generating point-based
clinical scores. This 5-step process gives users the flexibility of
customization (e.g., determining the final list of variables according
to the parsimony plot, and fine-tuning the cutoffs in variable
transformation). Please follow the step-by-step instructions (in Demos)
to build your own scores.
- STEP(i):
AutoScore_Survival_rank()- Rank variables with Random Survival Forest (AutoScore-Survival Module 1) - STEP(ii):
AutoScore_Survival_parsimony()- Select the best model with parsimony plot (iAUC) (AutoScore-Survival Modules 2+3+4) - STEP(iii):
AutoScore_Survival_weighting()- Generate the initial score with the final list of variables (Re-run AutoScore-Survival Modules 2+3) - STEP(iv):
AutoScore_Survival_fine_tuning()- Fine-tune the score by revisingcut_vecwith domain knowledge (AutoScore-Survival Module 5) - STEP(v):
AutoScore_Survival_testing()- Evaluate the final score with ROC(t) analysis (AutoScore-Survival Module 6)
Note: This is just the initial version of the AutoScore-Survival. Further version will be developed and updated.
Xie F, Ning Y, Yuan H, Goldstein BA, Ong MEH, Liu N, Chakraborty B. AutoScore-Survival: developing interpretable machine learning-based time-to-event scores with right-censored survival data. arXiv:2106.06957 (https://arxiv.org/abs/2106.06957)
Xie F, Chakraborty B, Ong MEH, Goldstein BA, Liu N. AutoScore: A Machine Learning-Based Automatic Clinical Score Generator and Its Application to Mortality Prediction Using Electronic Health Records. JMIR Medical Informatics 2020;8(10):e21798 (http://dx.doi.org/10.2196/21798)
- Feng Xie (Email: xief@u.duke.nus.edu)
- Nan Liu (Email: liu.nan@duke-nus.edu.sg)
library(survival)
library(randomForestSRC)
library(survAUC)
library(knitr)
source('D:/Document/AutoScore-Survival/R/AutoScore_Survival.R')
load("D:/Document/sample_data_survival.RData")- Read data from CSV or Excel files.
sample_data_survivalhas 10000 samples, with the same distribution as the data in the MIMIC-III ICU database (https://mimic.mit.edu/).
head(sample_data_survival)
#> Age GENDER ETHNICITY INSURANCE heartrate_mean sysbp_mean diasbp_mean
#> 13515 59.39907 F Others Private 83.77419 106.6364 43.12121
#> 18738 46.66896 M WHITE Medicaid 66.44000 124.5217 76.13043
#> 3413 81.67858 M WHITE Medicare 89.61111 121.9394 63.18182
#> 36336 68.24655 M WHITE Medicare 97.11111 142.6429 64.78571
#> 16682 39.93715 F Others Medicaid 114.03226 138.4348 86.34783
#> 57488 75.56852 M WHITE Medicare 84.23077 108.6250 61.04167
#> meanbp_mean resprate_mean tempc_mean spo2_mean glucose_mean aniongap_mean
#> 13515 67.19192 17.89189 36.84286 99.33333 137.7692 11.0
#> 18738 88.17391 14.96000 36.27778 97.12000 96.0000 12.5
#> 3413 81.90909 17.29545 36.73810 97.62857 226.4444 13.0
#> 36336 89.70238 18.14815 37.04365 96.55556 160.5714 23.0
#> 16682 98.18182 25.55556 37.45555 98.53125 76.0000 12.0
#> 57488 72.12500 22.30769 37.42857 97.73077 138.7500 11.5
#> bicarbonate_mean creatinine_mean chloride_mean lactate_mean
#> 13515 27.0 0.40 102.0 1.8
#> 18738 26.5 1.00 107.0 1.8
#> 3413 25.0 1.10 116.0 2.4
#> 36336 20.5 10.40 95.5 2.1
#> 16682 26.0 1.10 107.0 3.6
#> 57488 23.5 0.55 101.0 1.8
#> hemoglobin_mean hematocrit_mean platelet_mean potassium_mean bun_mean
#> 13515 10.60 32.00 179.0 4.45 9.0
#> 18738 13.05 38.35 300.0 4.45 22.5
#> 3413 12.20 35.00 110.0 3.90 17.0
#> 36336 7.80 24.35 174.0 4.40 106.5
#> 16682 12.70 37.55 276.0 4.15 10.5
#> 57488 9.55 28.25 404.5 3.80 9.5
#> sodium_mean wbc_mean time status
#> 13515 136.5 15.10 91 0
#> 18738 141.5 4.95 91 0
#> 3413 150.0 10.40 6 1
#> 36336 134.5 15.70 73 1
#> 16682 140.5 8.00 91 0
#> 57488 132.0 12.20 23 1- Handle missing values (AutoScore-Survival requires a complete dataset).
- Remove special characters from variable names, e.g.,
[,],(,),,. (Suggest using_to replace them if needed) - Name of the variable should be unique and not entirely included by other variable names.
- Ensure that there are dependent variables (“time” and “status”)
- Independent variables should be numeric (class: num/int) or categorical (class: factor/logic).
- Handle outliers (optional).
- Check variable distribution (optional).
- Check if data fulfill the basic requirement by AutoScore-Survival.
- Fix the problem if you see any warnings.
check_data(sample_data_survival)
#>
#> missing value check passed.- Modify your data, and run the
check_dataagain until there are no warning messages.
In Demo #1, we demonstrate the use of AutoScore-Survival on a comparably large dataset where separate training and validation sets are available. Please note that it is just a demo using simulated data, and thus, the result might not be clinically meaningful.
- Option 1: Prepare three separate datasets to train, validate, and test models.
- Option 2: Use demo codes below to randomly split your dataset into training, validation, and test datasets (70%, 10%, 20%, respectively).
set.seed(4)
out_split <- split_data(data = sample_data_survival, ratio = c(0.7, 0.1, 0.2))
train_set <- out_split$train_set
validation_set <- out_split$validation_set
test_set <- out_split$test_setntree: Number of trees in the random forest algorithm (Default: 100).
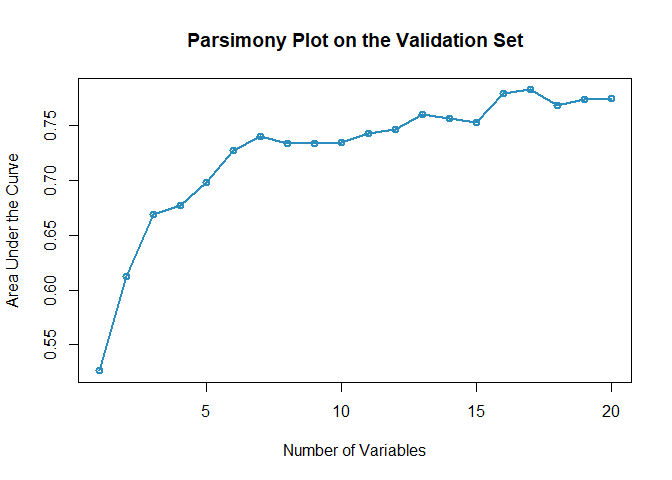
ranking <- AutoScore_Survival_rank(train_set, ntree = 100)nmin: Minimum number of selected variables (Default: 1).nmax: Maximum number of selected variables (Default: 20).categorize: Methods for categorizing continuous variables. Options include"quantile"or"kmeans"(Default:"quantile").quantiles: Predefined quantiles to convert continuous variables to categorical ones. (Default:c(0, 0.05, 0.2, 0.8, 0.95, 1)) Available ifcategorize = "quantile".max_cluster: The max number of cluster (Default: 5). Available ifcategorize = "kmeans".max_score: Maximum total score (Default: 100).
AUC <- AutoScore_Survival_parsimony(
train_set,
validation_set,
rank = ranking,
max_score = 100,
n_min = 1,
n_max = 20,
categorize = "quantile",
quantiles = c(0, 0.05, 0.2, 0.8, 0.95, 1)
)
#> Select 1 Variable(s): 0.5264094
#> Select 2 Variable(s): 0.6125118
#> Select 3 Variable(s): 0.6685712
#> Select 4 Variable(s): 0.6771743
#> Select 5 Variable(s): 0.6977727
#> Select 6 Variable(s): 0.7275343
#> Select 7 Variable(s): 0.7397659
#> Select 8 Variable(s): 0.7334412
#> Select 9 Variable(s): 0.7332976
#> Select 10 Variable(s): 0.7341622
#> Select 11 Variable(s): 0.7431479
#> Select 12 Variable(s): 0.7463571
#> Select 13 Variable(s): 0.759708
#> Select 14 Variable(s): 0.7563187
#> Select 15 Variable(s): 0.7530841
#> Select 16 Variable(s): 0.7788929
#> Select 17 Variable(s): 0.782435
#> Select 18 Variable(s): 0.7678376
#> Select 19 Variable(s): 0.7741454
#> Select 20 Variable(s): 0.7750084- Users could use the
AUCfor further analysis or export it as the CSV to other software for plotting.
write.csv(data.frame(AUC), file = "D:/AUC.csv")- Determine the optimal number of variables (
num_var) based on the parsimony plot obtained in STEP(ii). - The final list of variables is the first
num_varvariables in the ranked listrankingobtained in STEP(i). - Optional: User can adjust the finally included variables
final_variablesbased on the clinical preferences and knowledge.
# Example 1: Top 6 variables are selected
num_var <- 6
final_variables <- names(ranking[1:num_var])
# Example 2: Top 9 variables are selected
num_var <- 9
final_variables <- names(ranking[1:num_var])
# Example 3: Top 6 variables, the 9th and 10th variable are selected
num_var <- 6
final_variables <- names(ranking[c(1:num_var, 9, 10)])STEP(iii): Generate initial scores with the final list of variables (Re-run AutoScore-Survival Modules 2+3)
- Generate
cut_vecwith current cutoffs of continuous variables, which can be fine-tuned in STEP(iv). time_point: The time points to be evaluated using time-dependent AUC(t).
cut_vec <- AutoScore_Survival_weighting(
train_set,
validation_set,
final_variables,
max_score = 100,
categorize = "quantile",
quantiles = c(0, 0.05, 0.2, 0.8, 0.95, 1),
time_point = c(1,3,7,14,30,60,90)
)
#> ****Included Variables:
#> variable_name
#> 1 Age
#> 2 bun_mean
#> 3 resprate_mean
#> 4 creatinine_mean
#> 5 aniongap_mean
#> 6 lactate_mean
#> ****Initial Scores:
#>
#>
#> =============== =========== =====
#> variable interval point
#> =============== =========== =====
#> Age <30 0
#> [30,48.8) 11
#> [48.8,77.9) 15
#> [77.9,85.2) 22
#> >=85.2 24
#>
#> bun_mean <7.5 0
#> [7.5,11.5) 4
#> [11.5,34) 10
#> [34,66) 21
#> >=66 25
#>
#> resprate_mean <13.3 4
#> [13.3,15.4) 0
#> [15.4,21.3) 3
#> [21.3,25.6) 8
#> >=25.6 12
#>
#> creatinine_mean <0.5 17
#> [0.5,0.7) 7
#> [0.7,1.6) 2
#> [1.6,4.4) 4
#> >=4.4 0
#>
#> aniongap_mean <9.5 0
#> [9.5,11.5) 2
#> [11.5,16.5) 3
#> [16.5,20.5) 4
#> >=20.5 8
#>
#> lactate_mean <1 0
#> [1,1.5) 0
#> [1.5,2.3) 0
#> [2.3,4.3) 4
#> >=4.3 15
#> =============== =========== =====
#> Integrated AUC by all time points: 0.7275343
#> C_index: 0.711777
#> The AUC(t) are shown as bwlow:
#> time_point AUC_t
#> 1 1 0.7629788
#> 2 3 0.7032028
#> 3 7 0.7277124
#> 4 14 0.7207447
#> 5 30 0.7240616
#> 6 60 0.7413147
#> 7 90 0.7299956
#> ***The cutoffs of each variable generated by the AutoScore are saved in cut_vec. You can decide whether to revise or fine-tune themSTEP(iv): Fine-tune the initial score generated in STEP(iii) (AutoScore-Survival Module 5 & Re-run AutoScore-Survival Modules 2+3)
- Revise
cut_vecwith domain knowledge to update the scoring table (AutoScore-Survival Module 5). - Re-run AutoScore-Survival Modules 2+3 to generate the updated scores.
- Users can choose any cutoff values and/or any number of categories, but are suggested to choose numbers close to the automatically determined values.
## For example, we have current cutoffs of continuous variable: Age
## =============== =========== =====
## variable interval point
## =============== =========== =====
## Age <31.3 0
## [31.3,49.1) 12
## [49.1,78.2) 17
## [78.2,85.2) 22
## >=85.2 25 - Current cutoffs:
c(31.3, 49.1, 78.2, 85.2). We can fine tune the cutoffs as follows:
# Example 1: rounding up to a nice number
cut_vec$Age <- c(35, 50, 75, 85)
# Example 2: changing cutoffs according to clinical knowledge or preference
cut_vec$Age <- c(25, 50, 75, 85)
# Example 3: combining categories
cut_vec$Age <- c(50, 75, 85)- Then we do similar checks for other variables and update scoring table using new cutoffs if needed.
time_point: The time points to be evaluated using time-dependent AUC(t).
cut_vec$lactate_mean <- c(0.2, 1, 3, 4)
cut_vec$bun_mean <- c(10, 40)
cut_vec$aniongap_mean <- c(10, 17)
scoring_table <- AutoScore_Survival_fine_tuning(train_set,
validation_set,
final_variables,
cut_vec,
max_score = 100,
time_point = c(1,3,7,14,30,60,90))
#> ***Fine-tuned Scores:
#>
#>
#> =============== =========== =====
#> variable interval point
#> =============== =========== =====
#> Age <50 0
#> [50,75) 7
#> [75,85) 16
#> >=85 19
#>
#> bun_mean <10 0
#> [10,40) 12
#> >=40 25
#>
#> resprate_mean <13.3 6
#> [13.3,15.4) 0
#> [15.4,21.3) 4
#> [21.3,25.6) 10
#> >=25.6 15
#>
#> creatinine_mean <0.5 17
#> [0.5,0.7) 5
#> [0.7,1.6) 0
#> [1.6,4.4) 5
#> >=4.4 1
#>
#> aniongap_mean <10 0
#> [10,17) 2
#> >=17 5
#>
#> lactate_mean <1 0
#> [1,3) 1
#> [3,4) 6
#> >=4 19
#> =============== =========== =====
#> ***Performance (based on validation set, after fine-tuning):
#> Integrated AUC by all time points: 0.6996139
#> C_index: 0.6830529
#> The AUC(t) are shown as bwlow:
#> time_point AUC_t
#> 1 1 0.7675151
#> 2 3 0.6779422
#> 3 7 0.7028370
#> 4 14 0.6927692
#> 5 30 0.6850160
#> 6 60 0.7051068
#> 7 90 0.6991369time_point: The time points to be evaluated using time-dependent AUC(t).with_label: Set toTRUEif there are labels in thetest_setand performance will be evaluated accordingly (Default:TRUE).- Set the
with_labeltoFALSEif there are notlabelin thetest_setand the final predicted scores will be the output without performance evaluation.
pred_score <- AutoScore_Survival_testing(test_set, final_variables, cut_vec, scoring_table, threshold = "best", with_label = TRUE, time_point = c(1,3,7,14,30,60,90))
#> ***Performance using AutoScore (based on unseen test Set):
#> Integrated AUC by all time points: 0.7325347
#> C_index: 0.7046158
#> The AUC(t) are shown as bwlow:
#> time_point AUC_t
#> 1 1 0.7359898
#> 2 3 0.7597619
#> 3 7 0.7432432
#> 4 14 0.7248715
#> 5 30 0.7167943
#> 6 60 0.7316393
#> 7 90 0.7208223
head(pred_score)
#> pred_score time status
#> 1 33 91 0
#> 2 38 91 0
#> 3 26 91 0
#> 4 35 91 0
#> 5 49 14 1
#> 6 26 91 0- Users could use the
pred_scorefor further analysis or export it as the CSV to other software.
write.csv(pred_score, file = "D:/pred_score.csv")