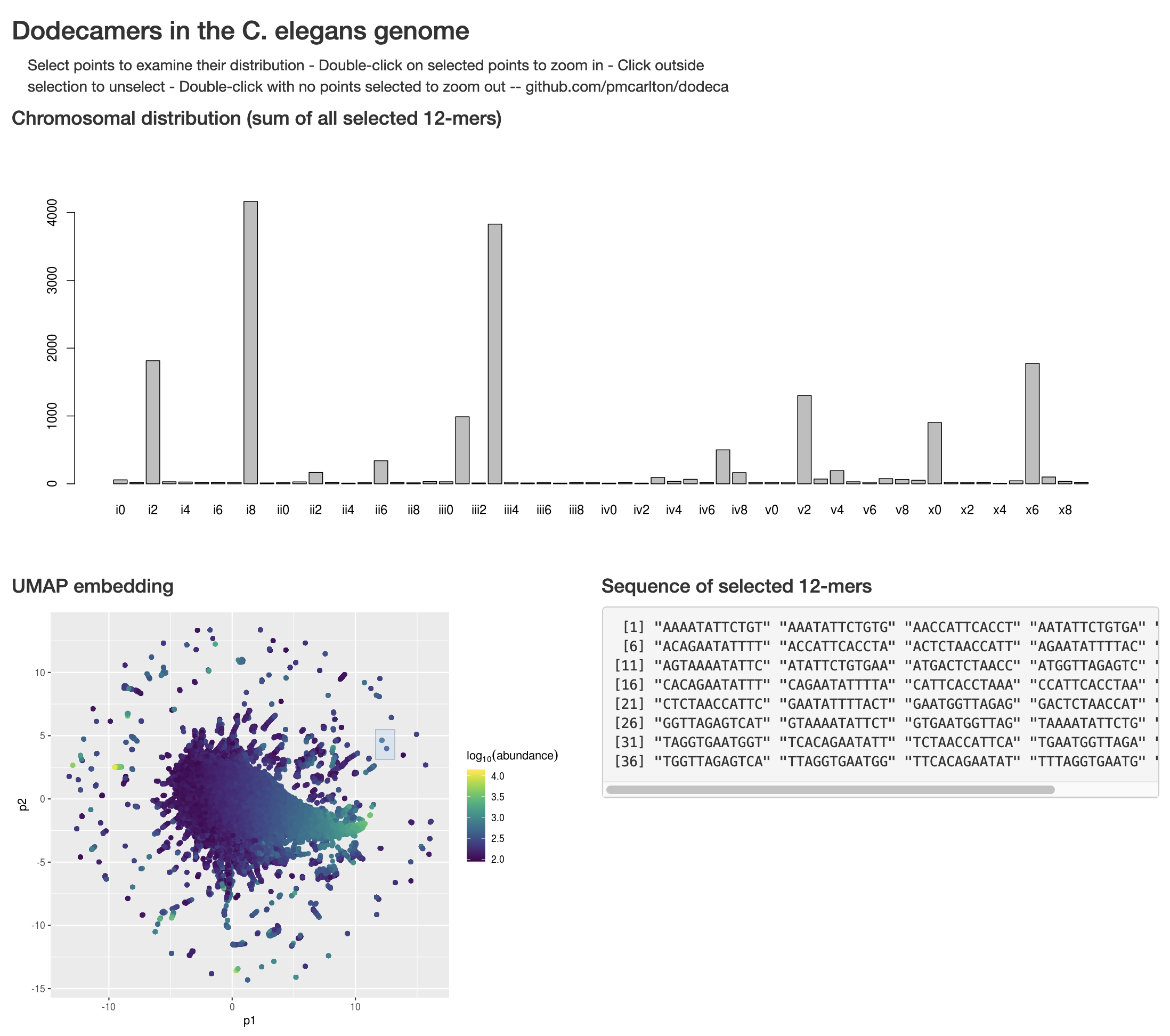
Dodecamer catalog by chromosome
Goal: Explore chromosome positions of all dodecamers in the C. elegans genome (current data from WS268)
Approach:
-
Using
mercatalog_multiple_hist.pl, calculate distribution of all 12-mers in bins of 10 along all 6 chromosomes. Details: read in each chromosome 12 bases at a time, and increment an array position inside a hash keyed by the current 12mer at the bin corresponding to the current position (i.e., each hash entry is keyed by a 12mer and contains an anonymous array that is a histogram of that 12mer's distribution) -
Create a UMAP embedding of the set of all dodecamers that occur >=100 times in the genome (using n_neighbors=25, min_dist=0.2) (data in the .Rdat file shows sequence, histograms, and UMAP coordinates)
-
Display the embedding as an interactive webpage using Shiny, currently living at ilas.carltonlab.org/shiny/dodeca/ - warning, takes ~15 seconds to load and display anything)