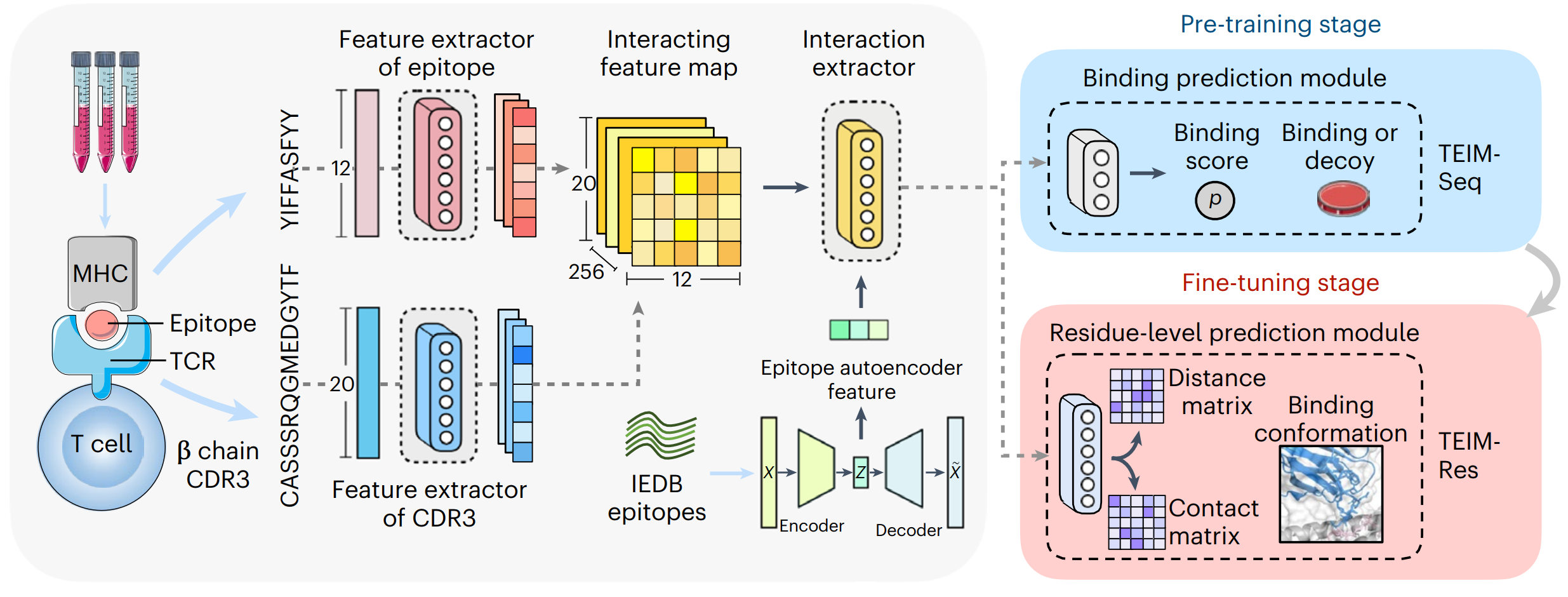
This repository contains the codes for the paper TEIM: Characterizing the interaction conformation between T-cell receptors and epitopes with deep learning published in Nature Machine Intelligence.
TEIM (TCR-Epitope Interaction Modeling) is a deep learning-based model to predict the TCR-epitope interactions, including two submodels TEIM-Res (TEIM at Residue level) and TEIM-Samp (TEIM at Sequence level).
Both models only takes the primary sequences of CDR3βs and the epitopes as input. TEIM-Res predicts the distances and the contact probabilities between all residue pairs of CDR3βs and epitopes. TEIM-Seq predicts whether the CDR3βs and epitopes can bind to each other.
- Install Python>=3.8 and Anaconda.
- Install basic packages using:
Note: Change the Pytorch version to be compatible with your CUDA version. Besides, since the Pytorch Lightning version we used is 1.6.4, the compatible Pytorch version is
# [Optional] Create a new environment and activate it conda create -n teim python=3.8 conda activate teim # Install Pytorch packages (for CUDA 11.3) conda install pytorch==1.10.1 cudatoolkit=11.3 -c pytorch -c conda-forge # Install other packages pip install -r requirements.txt$>=1.8,<=1.11$ (see here). - Install ANARCI for CDR3 numbering.
conda install -c bioconda anarci
We also provided a docker file to facilitate the installation of environment. You can build the docker by runing
docker build -t teim:v1 .-
Put your input TCR-epitope sequence pairs in the
inputs/inputs.csvfile. The TCRs are represented by their CDR3β sequences and the epitopes are represented by their sequences in the following format:cdr3 epitope CASAPGLAGGRPEQYF LLFGYPVYV CASRGAAGGRPQYF MLWGYLQYV CASRPGLAGGRAEQYF FTDSSVWA -
Run
python scripts/inference_res.py -
The predicted distance matrices and contact site matrices are in the
outputsdirectory:- The predicted distance matrix and contact matrix are in the files names as
dist_<cdr3>_<epitope>.csvandsite_<cdr3>_<epitope>.csv, respectively. - The rows and columns of the matrices represent the CDR3βs and epitopes, respectively.
- The values in the distance matrix stand for the distances of residue pairs (unit: angstrom) and the values in the contact matrix stand for the predicted contact scores (probabilities) of residue pairs (range from 0 to 1).
- The predicted distance matrix and contact matrix are in the files names as
- Put your input TCR-epitope sequence pairs in the
inputs/inputs_bd.csvfile. The format is the same asinputs/inputs.csv(residue-level input file). - Run
python scripts/inference_seq.py - The predicted sequence-level binding scores are in the
outputs/sequence_level_binding.csv. Thebindingcolumn in the file represent the predicted sequence-level binding scores (probabilities) of the TCR-epitope pair.
Please refer to the directory train_teim.
@article{Peng2023,
doi = {10.1038/s42256-023-00634-4},
url = {https://doi.org/10.1038/s42256-023-00634-4},
year = {2023},
month = mar,
publisher = {Springer Science and Business Media {LLC}},
volume = {5},
number = {4},
pages = {395--407},
author = {Xingang Peng and Yipin Lei and Peiyuan Feng and Lemei Jia and Jianzhu Ma and Dan Zhao and Jianyang Zeng},
title = {Characterizing the interaction conformation between T-cell receptors and epitopes with deep learning},
journal = {Nature Machine Intelligence}
}If you have any questions, please contact us at xingang.peng@gmail.com