Robust comparative analysis and contamination removal for metagenomic data
With Recentrifuge, researchers can interactively explore what organisms are in their samples and at which level of confidence, thus enabling a robust comparative analysis of multiple samples in any metagenomic study.
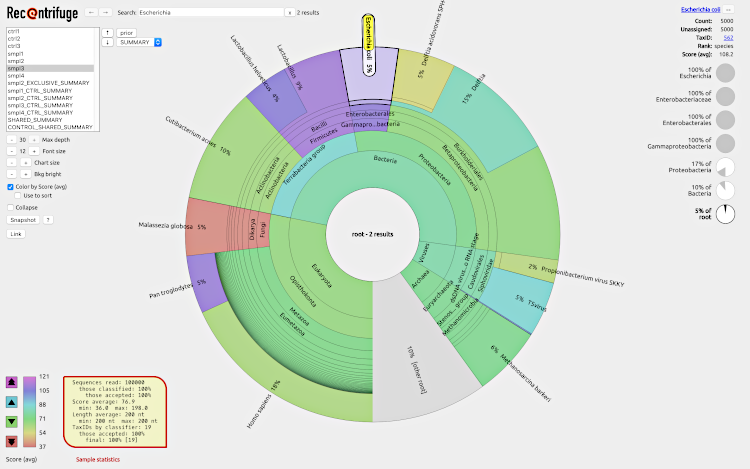
Recentrifuge enables researchers to analyze results from the Centrifuge and LMAT classifiers using interactive pie charts, by placing great emphasis on the confidence level (score) of the taxonomic classifications.
In addition to the scored charts for the original samples, different sets of scored plots are generated for each taxonomic level of interest, like shared taxa and exclusive taxa per sample.
If there are one or more control samples in the study, Recentrifuge will also generate control-subtracted plots and a shared taxa plot with the controls removed. The robust control removal algorithm detects and selectively removes various types of contamination, including crossovers.
Recentrifuge is especially useful in the case of low biomass metagenomic studies and when a more reliable detection of minority organisms is needed, like in clinical, environmental and forensic analysis.
For usage and documentation, please, see the Recentrifuge wiki.
Further details in the bioRxiv pre-print.
To see an example of webpage generated by Recentrifuge, click on the screenshot: