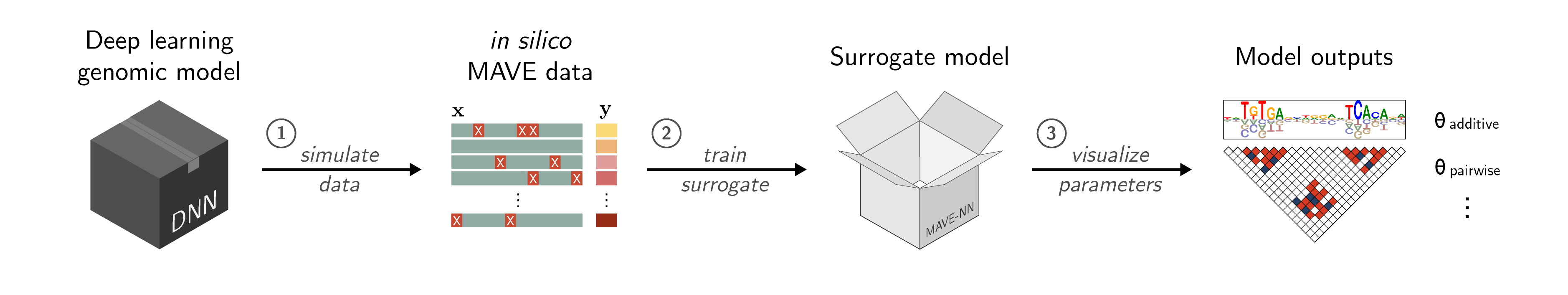
SQUID (Surrogate Quantitative Interpretability for Deepnets) is a Python suite to interpret sequence-based deep learning models for regulatory genomics data with domain-specific surrogate models. For installation instructions, tutorials, and documentation, please refer to the SQUID website, https://squid-nn.readthedocs.io/. For an extended discussion of this approach and its applications, please refer to our manuscript:
- Seitz E., McCandlish D., Kinney J., and Koo P.K. Identifying cis-regulatory mechanisms from genomic deep neural networks with surrogate models.
pip install squid-nnpip install logomaker
pip install mavenn --upgradeFor issues installing MAVE-NN, please see:
For older DNNs that require inference via Tensorflow 1.x, Python 2.x is required which is not supported by MAVE-NN. Users will need to create separate environments in this case:
- Tensorflow 1.x and Python 2.x environment for generating in silico MAVE data
- Tensorflow 2.x and Python 3.x environment for training MAVE-NN surrogate models
SQUID provides a simple interface that takes as input a sequence-based deep-learning model (e.g., a DNN), which is used as an oracle to generate an in silico MAVE dataset representing a localized region of sequence space. The MAVE dataset can then be fit using a domain-specific surrogate model, with the resulting parameters visualized to reveal the cis-regulatory mechanisms driving model performance.
-
Examples in Google Colab:
-
Additive analysis with DeepSTARR: https://colab.research.google.com/drive/12HR8Vu_8ji3Ac1wli4wgqx1J0YB73JF_?usp=sharing
-
Pairwise analysis with ResidualBind-32: https://colab.research.google.com/drive/1eKC78YE2l49mQFOlnA9Xr1Y9IO121Va5?usp=sharing
-
Variant effect analysis with Kipoi model: https://colab.research.google.com/drive/1wtpT1FF5nu1etTDOaV3A7ByXhuLqK071?usp=sharing
-
-
Examples in Python:
- The
squid-manuscriptrepository contains examples to reproduce results in the manuscript, including the application of SQUID on other DNNs such as ENFORMER: https://github.com/evanseitz/squid-manuscript
- The
If this code is useful in your work, please cite our paper.
@article{seitz2023_squid,
title={Identifying *cis*-regulatory mechanisms from genomic deep neural networks using surrogate models},
author={Seitz, Evan and McCandlish, David and Kinney, Justin and Koo, Peter},
journal={TBD},
volume={TBD},
number={TBD},
pages={TBD},
year={2023},
publisher={TBD}
}Copyright (C) 2022–2023 Evan Seitz, David McCandlish, Justin Kinney, Peter Koo
The software, code sample and their documentation made available on this website could include technical or other mistakes, inaccuracies or typographical errors. We may make changes to the software or documentation made available on its web site at any time without prior notice. We assume no responsibility for errors or omissions in the software or documentation available from its web site. For further details, please see the LICENSE file.