CREME (get the money!)
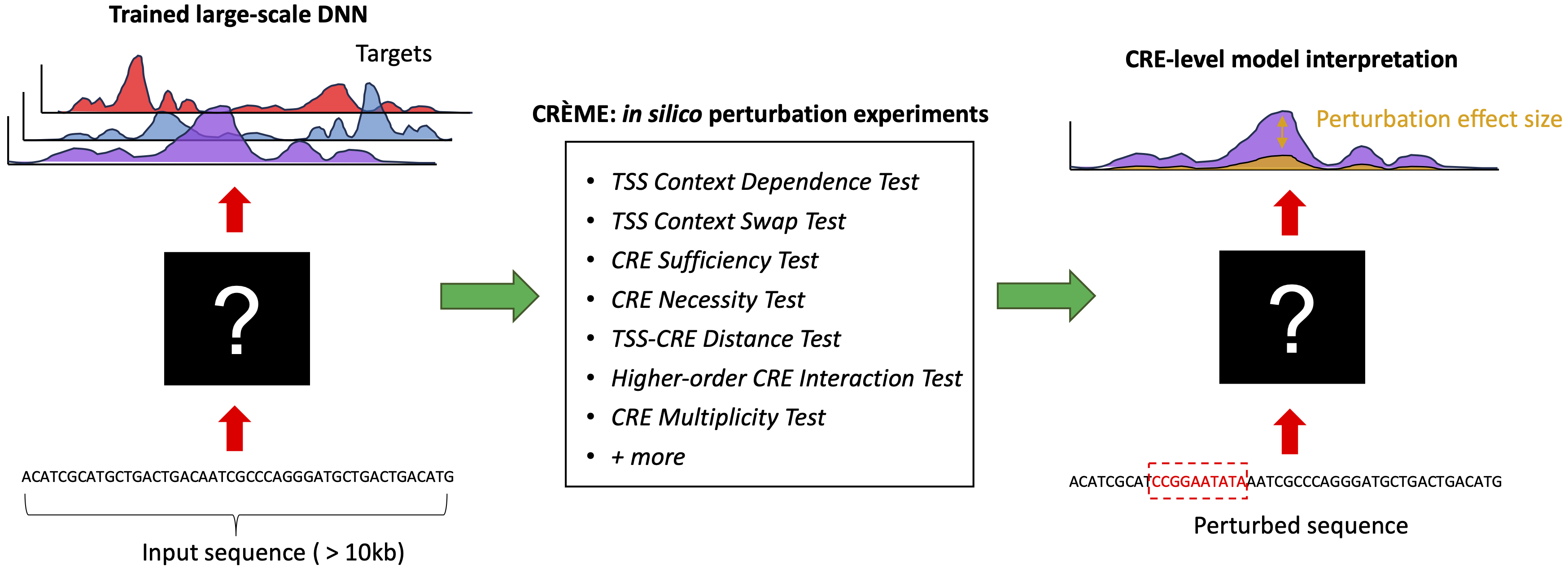
CREME (Cis-Regulatory Element Model Explanations) is an in silico perturbation framework designed to examine large-scale DNNs trained on regulatory genomics data. CREME can provide interpretations at various scales, including a coarse-grained CRE-level view as well as a fine-grained motif-level view. CREME can be used to identify cis-regulatory elements (CREs) that directly enhance or silence target genes. CREME can also be used to map CRE distance from transcription start sites and gene expression, as well as the intricate complexity of higher-order CRE interactions.
CREME is based on the notion that by fitting experimental data, the DNN essentially approximates the underlying "function" of the experimental assay. Thus, the trained DNN can be treated as a surrogate for the experimental assay, enabling in silico "measurements" for any sequence. CREME comprises a suite of perturbation experiments to uncover how DNNs learn rules of interactions between CREs and their target genes
Paper: https://www.biorxiv.org/content/10.1101/2023.07.03.547592v1
Tutorial and full documentation on Readthedocs.org comding soon...
CREME is pip installable:
pip install creme-nn