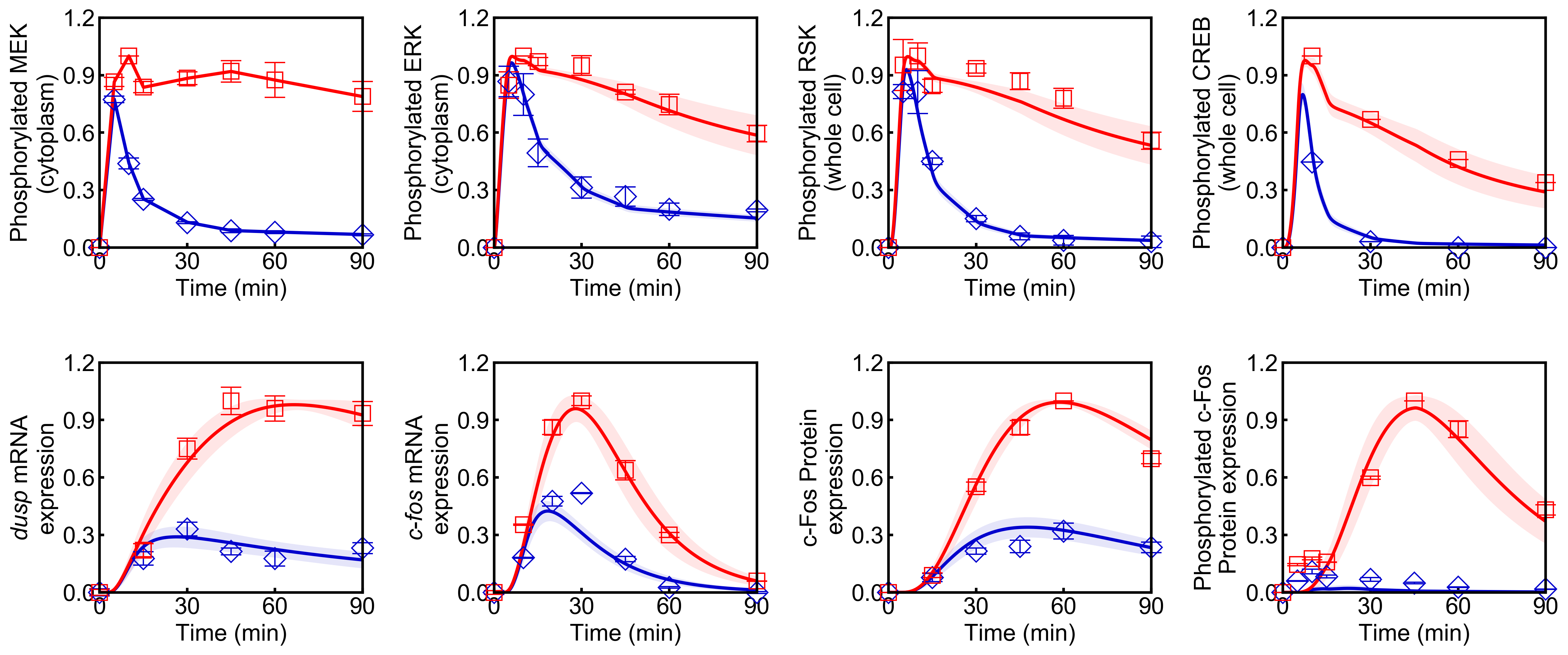
Using Genetic Algorithm to Fit ODE Models to Data
Points (blue diamonds, EGF; red squares, HRG) denote experimental data, solid lines denote simulations.
- numpy
- scipy
- matplotlib
- seaborn
- jupyter
A brief description of each file you will need to use is below:
-
Name Description name2idx/This is where you define the parameter/variable names of your model. set_model.pyThis file contains the differential equations, parameters and initial concentrations. observalbe.pyThis is the file to define the simulations you want to run and input the experimental data that you are going to use to try and fit the parameters to. set_search_param.pyThis is where you specify the model parameters to estimate. fitness.pyThis is where you define an objective function to minimize, e.g. the distance between model simulation and experimental data. -
runGA_n.ipynb (n ≧ 1)
Run both input boxes and leave it. When you run the ith file, runGA_(i+1).ipynb will be generated. You can run these different parameter fittings simultaneously. -
runSim.ipynb
This is the file that is used to actually run the simulations for your model and plot the results. -
figure/
Within this folder there are several figures that will be saved. One is the ‘param_range.pdf’ which shows the range of values for your parameters based on all the fittings. The others are results of your simulations.
%%javascript
IPython.notebook.kernel.execute(
'current_ipynb = "' + IPython.notebook.notebook_name + '"'
)%run -i optimize.py
""" If you want to continue from where you stopped in the last parameter search,
%run -i optimize_continue.py
"""or
$ nohup python optimize.py n &- If you want to search multiple parameter sets (from n1 to n2) simultaneously,
$ nohup python optimize.py n1 n2 &viz_type : str
-
'average': The average of simulation results with parameter sets infitparam/ -
'best': The best simulation result infitparam/, simulation withbest_fit_param -
'original': Simulation with the default parameters and initial values defined inmodel/ -
'n(=1,2,...)': Use the parameter set infitparam/n/
show_all : bool
- Whether to show all simulation results.
stdev : bool
- If True, the standard deviation of simulated values will be shown (only when
viz_type == 'average').
from param_estim import simulate_all
simulate_all(viz_type='average',show_all=False,stdev=True)$ git clone https://github.com/okadalabipr/intro_to_param_estim.git
We thank Benjamin Roberts and Iosifina Sampson (University of Leeds) for helpful comments and discussions.
-
Nakakuki, T. et al. Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell 141, 884–896 (2010). https://doi.org/10.1016/j.cell.2010.03.054
-
Kimura, S., Ono, I., Kita, H. & Kobayashi, S. An extension of UNDX based on guidelines for designing crossover operators: proposition and evaluation of ENDX. Trans. Soc. Instrum. Control Eng. 36, 1162–1171 (2000). https://doi.org/10.9746/sicetr1965.36.1162
-
Kimura, S. & Konagaya, A. A Genetic Algorithm with Distance Independent Diversity Control for High Dimensional Function Optimization. J. Japanese Soc. Artif. Intell. 18, 193–202 (2003). https://doi.org/10.1527/tjsai.18.193
-
Kimura, S., Nakakuki, T., Kirita, S. & Okada, M. AGLSDC: A Genetic Local Search Suitable for Parallel Computation. SICE J. Control. Meas. Syst. Integr. 4, 105–113 (2012). https://doi.org/10.9746/jcmsi.4.105