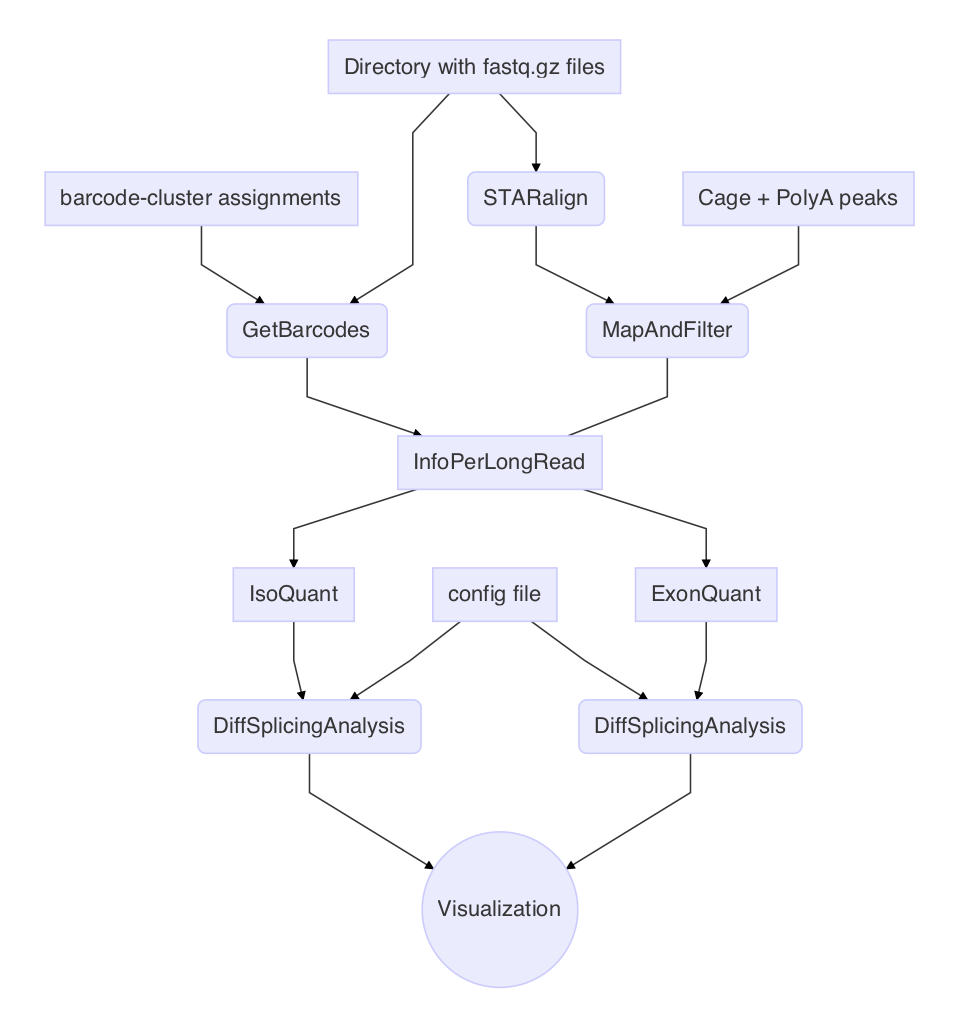
scisorseqr is a linux based R-package for analyzing differential isoform expression in single cells. The methods are based on our recent publication and the scISOrSeq workflow
Any comparative studies of alternative splicing can be performed with scisorseqr. The package includes functions for barcode deconvolution from fastqs, integration with long read alignment tools, mapping and filtering of high confidence, full-length spliced reads, and some handy tools to conduct differential expression analysis.
The tools are also applicable to long-read spatial transcriptomics, and can be used to resolve exon expression at the spatial level
The package has only been tested on a CentOS x86_64 machine. For alignment and mapping, we recommend
- STARlong software installation for PacBio reads
- Minimap2 installation for Oxford Nanopore (or PacBio) reads
- samtools
- bedtools
- python version 3.7
The easiest way to install scisorseqr is through Github with:
devtools::install_github('noush-joglekar/scisorseqr',build_vignettes = TRUE)These steps are available as functions in the package. For example, barcode deconvolution can be done using the following command
library(scisorseqr)
GetBarcodes('FastqFiles/','userInput/BarcodeCluster_Assignments', concatenate=TRUE,
filterReads=FALSE, numProcesses=24)A step-by-step outline of the various functions is available as a vignette. To access it, run
browseVignettes("scisorseqr")We appreciate any and all inputs for improving scisorseqr. Feel free to send us an email or open an issue.