DESeq2Shiny: a web-based app based on the DESeq2 R package for RNA-seq counts data exploratory analysis and differential expression
This Shiny app is a wrapper around DESeq2, an R package for "Differential gene expression analysis based on the negative binomial distribution".
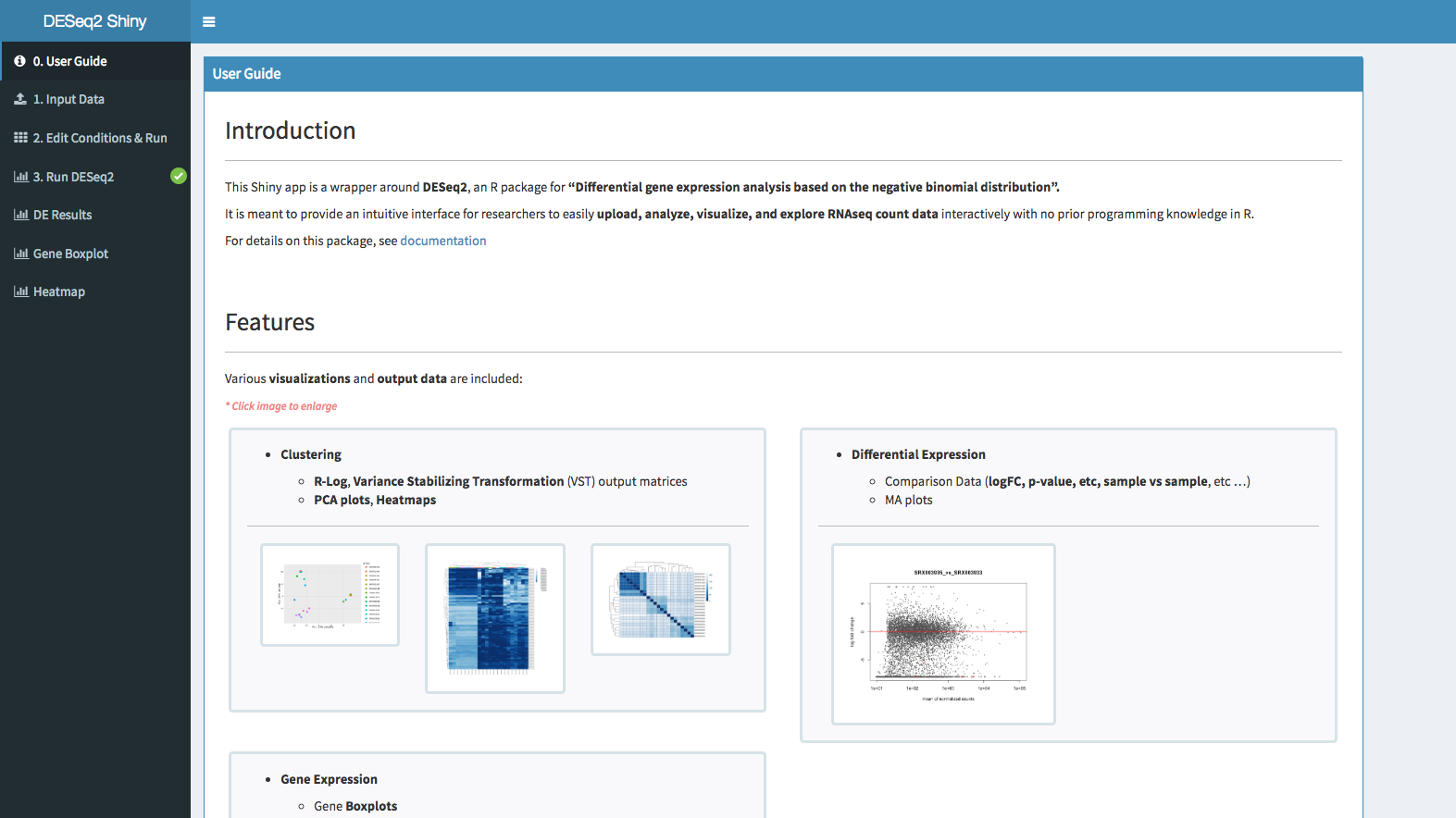
It is meant to provide an intuitive interface for researchers to easily upload, analyze, visualize, and explore RNAseq count data interactively with no prior programming knowledge in R.
This tool supports simple or multi-factorial experimental design. It also allows for exploratory analysis when no replicates are available.
The app also provides svaseq Surrogate Variable Analysis for hidden batch effect detection. The user can then include Surrogate Variables (SVs) as adjustment factors for downstream analysis (eg. differential expression). For more information on svaseq, go to this link
You can try it online at http://nasqar.abudhabi.nyu.edu/deseq2shiny
NASQAR: A web-based platform for High-throughput sequencing data analysis and visualization
Various visualizations and output data are included:
-
Clustering
- R-Log, Variance Stabilizing Transformation (VST) output matrices
- PCA plots, Heatmaps
-
Differential Expression
- Comparison Data (logFC, p-value, etc, sample vs sample, etc …)
- MA plots
-
Gene Expression
- Gene Boxplots