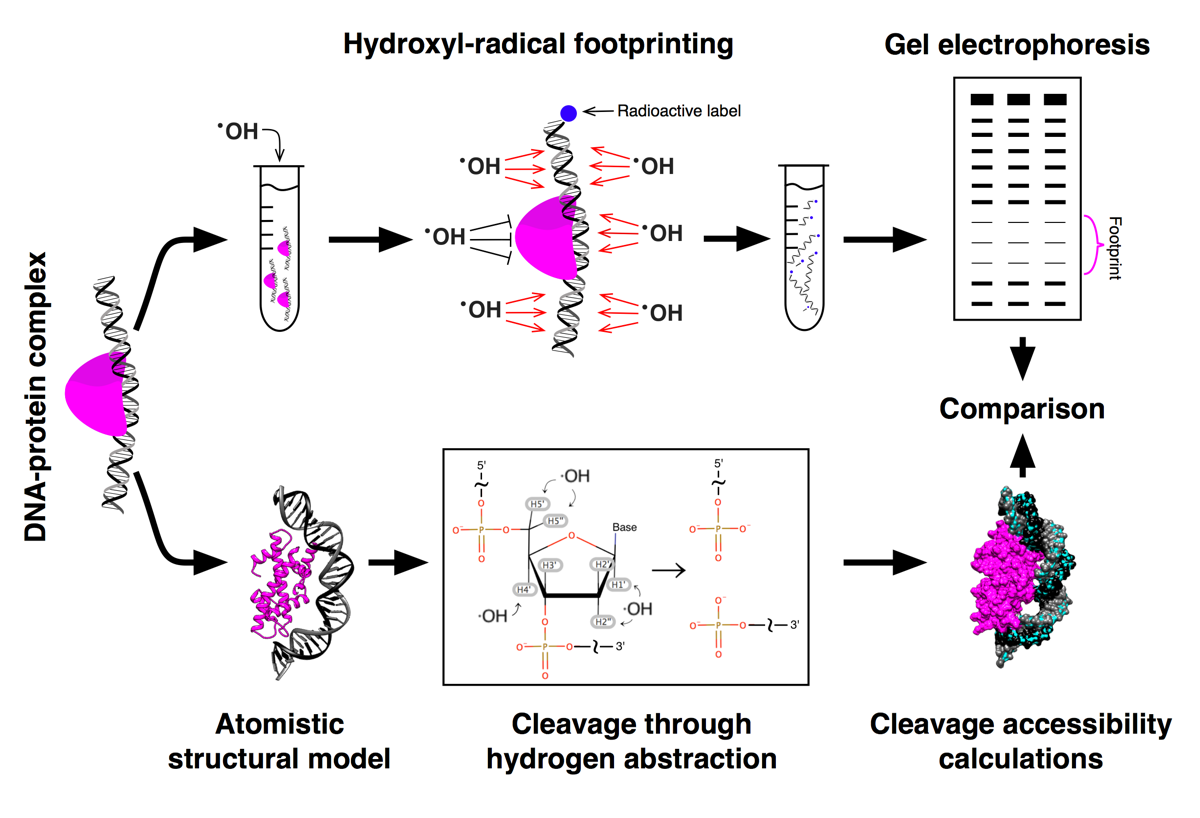
HYDROID (HYDroxyl-Radical fOotprinting Interpretation for DNA) is a python package for the analysis of the experimental data generated by hydroxyl-radical footprinting (HRF) of DNA-protein complexes and its interpretation through comparison to theoretical predictions from molecular models.
- 2 in 1: HRF experimental data quantification + theoretical analysis of atomistic structures
- Extracts cleavage intensities at individual DNA nucleotides by a number of constraint fitting algorithms
- Uses both Gaussian and Lorentzian models for band intensities
- Cross-platform python-scripted solution, can be install on Linux, MacOS, Windows
- Completely free and relies on open source components such as ImageJ and FreeSASA
- Provides examples of raw data analysis together with data analysis workflows.
For detailed documentation - click here.
Video tutorial is available here.
Install Miniconda with Python2.7 for your platform from https://conda.io/miniconda.html.
conda install -c hydroid hydroid
Test HYDROID:
HYDROID_test_exp #Tests exeprimental data analysis module
HYDROID_test_pred #Tests molecular structure analysis module (currently supported on Linux and OSX)
For alternative installation instructions for Linux, MacOS and PC see INSTALL.md.
HYDROID_get_ex1
cd example1
python exp_s2_assign_peaks.py
...
See full examples set and instructions in examples folder.
Please cite HYDROID using following publications:
- A.K. Shaytan et al. (in preparation)