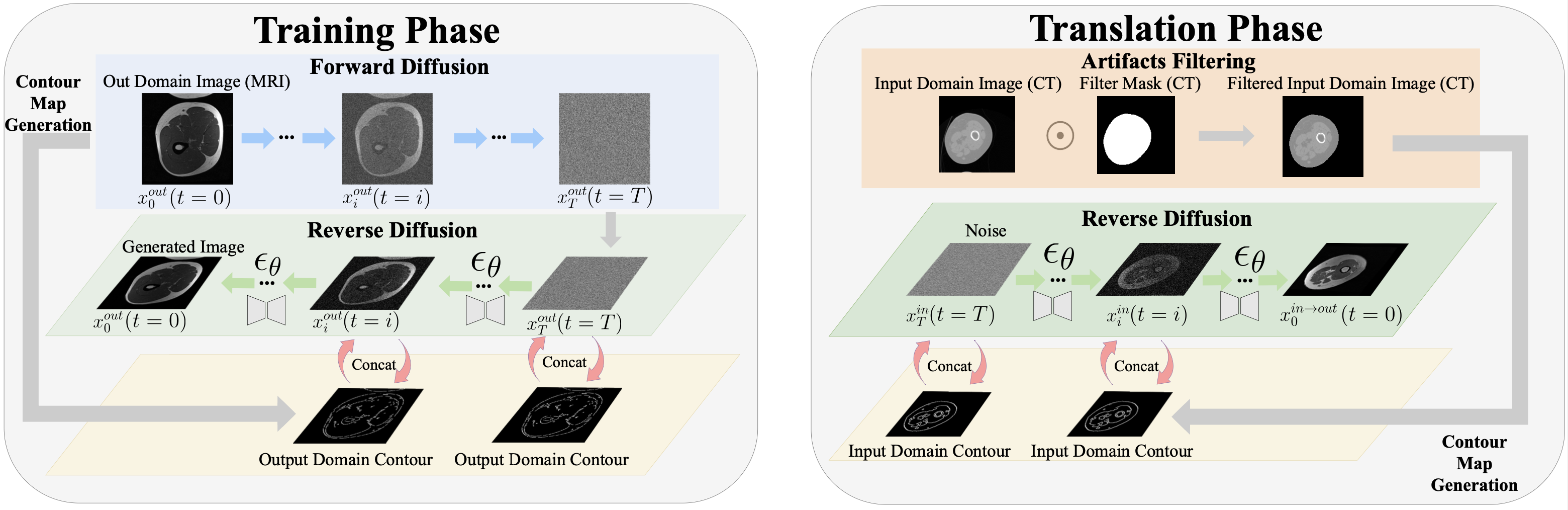
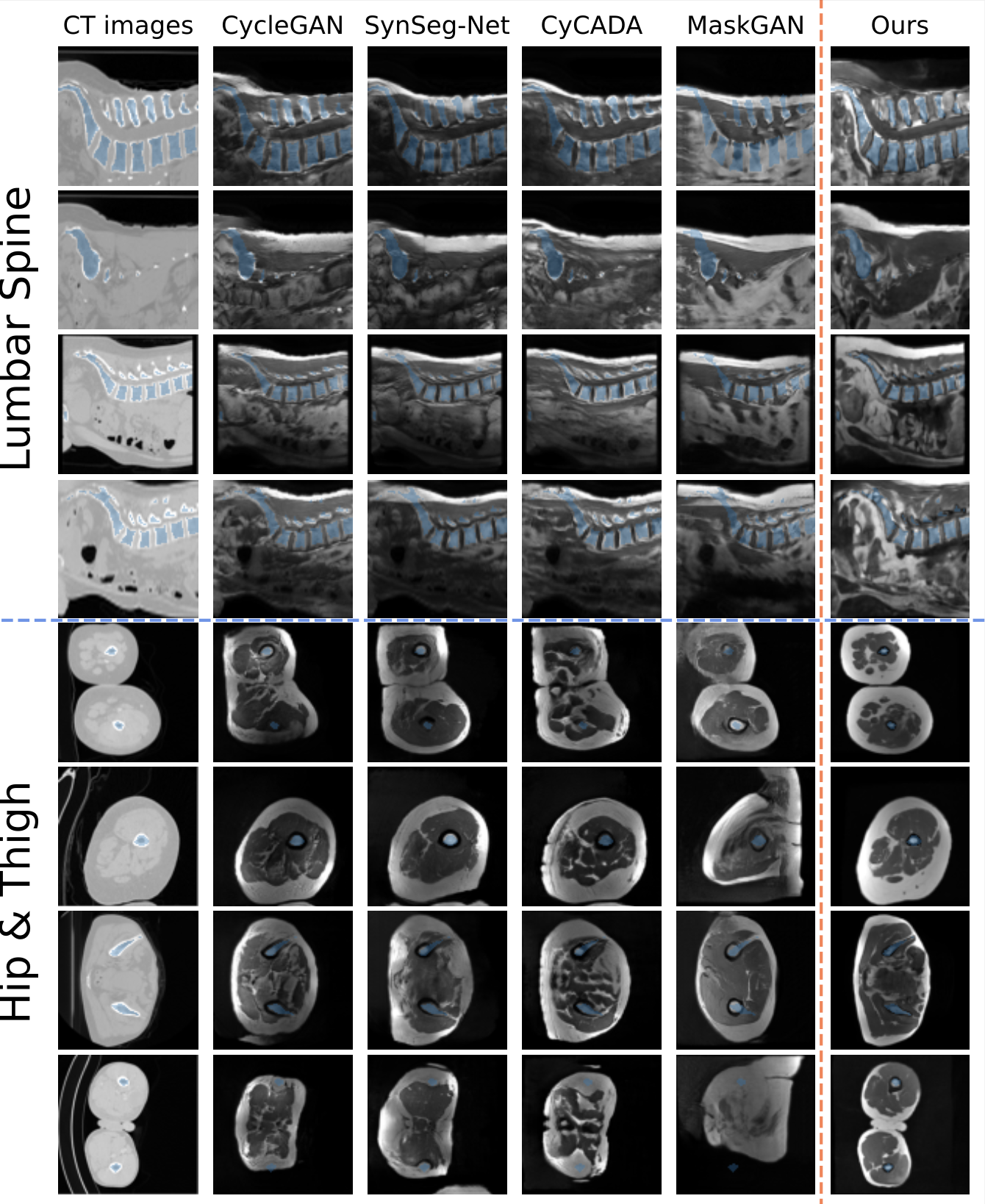
Contour-Guided Diffusion Models for Unpaired Image-to-Image Translation
By Yuwen Chen, Nicholas Konz, Hanxue Gu, Haoyu Dong, Yaqian Chen, Lin Li, Jisoo Lee and Maciej Mazurowski
This is the code for our paper ContourDiff: Unpaired Image Translation with Contour-Guided Diffusion Models, which is a novel framework that leverages domain-invariant anatomical contour representations of images to enable unpaired translation between different domains.
Our method can:
- Enforce precise anatomical consistency even between modelaities with severe structural biases (See example figure below)
- Potentially translate images from arbitrary unseen input domains (i.e., train once, translate any)
Great thanks to Segmentation-guided Diffusion for inspiration and code backbone!
Please cite our paper if you use our code or reference our work:
@article{chen2024contourdiff,
title={ContourDiff: Unpaired Image Translation with Contour-Guided Diffusion Models},
author={Chen, Yuwen and Konz, Nicholas and Gu, Hanxue and Dong, Haoyu and Chen, Yaqian and Li, Lin and Lee, Jisoo and Mazurowski, Maciej A},
journal={arXiv preprint arXiv:2403.10786},
year={2024}
}Please follow the steps below to have your own ContourDiff model!
To extract the contours, run command:
python preprocess.py \
--data_directory {DATA_DIRECTORY} \
--domain_img_folder {DOMAIN_IMG_FOLDER} \
--domain_contour_folder {DOMAIN_CONTOUR_FOLDER} \
--domain_meta_path {DOMAIN_META_PATH} \where:
DATA_DIRECTORYis directory of data from multiple domainsDOMAIN_IMG_FOLDERis path to certain domain images underDATA_DIRECTORYDOMAIN_CONTOUR_FOLDERis path to save extracted contours underDATA_DIRECTORYDOMAIN_META_PATHis path (*.csv) to save meta information underDATA_DIRECTORY
To enable removal of non-anatomical background artifacts, use --remove_artifact.
For example, given data structure below:
DATA_DIRECTORY
├── domain_1
│ ├── images
│ │ ├── img_1.png
│ │ ├── img_2.png
│ │ └── ...
├── domain_2
│ ├── images
│ │ ├── img_1.png
│ │ ├── img_2.png
│ │ └── ...
└── ...
If extracting contours for images from domain 1, then set DOMAIN_IMG_FOLDER="domain_1/images"
Then, if setting DOMAIN_CONTOUR_FOLDER="domain_1/contours" and DATA_DIRECTORY="domain_1/df_meta.csv", the outcome data structure is:
DATA_DIRECTORY
├── domain_1
│ ├── images
│ │ ├── img_1.png
│ │ ├── img_2.png
│ │ └── ...
│ ├── contours
│ │ ├── img_1.png
│ │ ├── img_2.png
│ │ └── ...
│ ├── df_meta.csv
├── domain_2
│ ├── images
│ │ ├── img_1.png
│ │ ├── img_2.png
│ │ └── ...
└── ...
To visualize the extracted contours, run contour_checker.ipynb.
To train your own ContourDiff model, run command:
python train.py \
--input_domain {INPUT_DOMAIN} \
--output_domain {OUTPUT_DOMAIN} \
--data_directory {DATA_DIRECTORY} \
--input_domain_img_folder {INPUT_DOMAIN_IMG_FOLDER} \
--input_domain_contour_folder {INPUT_DOMAIN_CONTOUR_FOLDER} \
--output_domain_img_folder {OUTPUT_DOMAIN_IMG_FOLDER} \
--output_domain_contour_folder {OUTPUT_DOMAIN_CONTOUR_FOLDER} \
--input_domain_meta_path {INPUT_DOMAIN_META_PATH} \
--output_domain_meta_path {OUTPUT_DOMAIN_META_PATH} \
--output_dir {OUTPUT_DIR}
--device {DEVICE} \
--contour_guided \
where:
INPUT_DOMAINis the string name of the input domain (e.g. any, CT or MRI)OUTPUT_DOMAINis the string name of the output domain (e.g. CT or MRI)DATA_DIRECTORYis directory of data from multiple domainsINPUT_DOMAIN_IMG_FOLDERis path to input domain images underDATA_DIRECTORYINPUT_DOMAIN_CONTOUR_FOLDERis path to input domain contours underDATA_DIRECTORYOUTPUT_DOMAIN_IMG_FOLDERis path to output domain images underDATA_DIRECTORYOUTPUT_DOMAIN_CONTOUR_FOLDERis path to output domain contours underDATA_DIRECTORYINPUT_DOMAIN_META_PATHis path (*.csv) to input domain meta file underDATA_DIRECTORYOUTPUT_DOMAIN_META_PATHis path (*.csv) to output domain meta file underDATA_DIRECTORYOUTPUT_DIRis absolute path to save output results, including model checkpoints and visualization samplesDEVICEis GPU devicecontour_guidedis flag to enable contour-guided mode for diffusion models
Notice: Input domain images and contours are used for validation in the training phase.
To translate input domain images using your own ContourDiff model, run command:
python translation.py \
--input_domain {INPUT_DOMAIN} \
--output_domain {OUTPUT_DOMAIN} \
--data_directory {DATA_DIRECTORY} \
--input_domain_contour_folder {INPUT_DOMAIN_CONTOUR_FOLDER} \
--input_domain_meta_path {INPUT_DOMAIN_META_PATH} \
--num_copy {NUM_COPY} \
--by_volume \
--volume_specifier {VOLUME_SPECIFIER} \
--slice_specifier {SLICE_SPECIFIER} \
--selected_epoch {SELECTED_EPOCH} \
--translating_folder_name {TRANSLATING_FOLDER_NAME} \
--device {DEVICE} \
--num_partition {NUM_PARTITION} \
--partition {PARTITION}
where:
INPUT_DOMAINis the string name of the input domain (e.g. any, CT or MRI)OUTPUT_DOMAINis the string name of the output domain (e.g. CT or MRI)DATA_DIRECTORYis directory of data from multiple domainsINPUT_DOMAIN_CONTOUR_FOLDERis path to input domain contours underDATA_DIRECTORYINPUT_DOMAIN_META_PATHis path (*.csv) to input domain meta file underDATA_DIRECTORYOUTPUT_DIRis absolute path to save output results, including model checkpoints and visualization samplesNUM_COPYis the number of samples generated in each iterationby_volumeis flag to enable slice-by-slice generation within each volumeVOLUME_SPECIFIERis string of column to indicate each volume (e.g., "volume")SLICE_SPECIFIERis string of column to indicate slice number (e.g., "slice")SELECTED_EPOCHis epoch of the selected checkpoint to loadTRANSLATING_FOLDER_NAMEis absolute path to store the tranlsated imagesDEVICEis GPU deviceNUM_PARTITIONis total number of partition to split input domain units (either slices or volumes)PARTITIONis specified partition to translate
Notice:
VOLUME_SPECIFIERandSLICE_SPECIFIERare required to enableby_volumetranslation, which means the meta file should include corresponding columns.NUM_PARTITIONandPARTITIONare aimed for translation in parallel.PARTITIONis within range [0,NUM_PARTITION- 1].